| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,042,164 – 7,042,258 |
| Length | 94 |
| Max. P | 0.938936 |

| Location | 7,042,164 – 7,042,258 |
|---|---|
| Length | 94 |
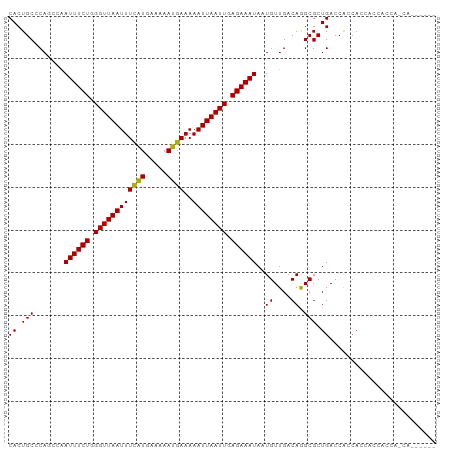
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Shannon entropy | 0.34360 |
| G+C content | 0.39483 |
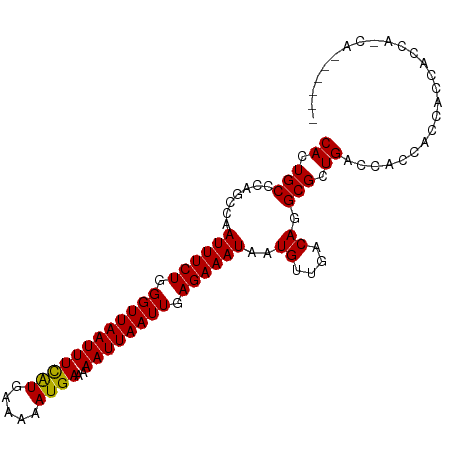
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -14.08 |
| Energy contribution | -13.58 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7042164 94 - 22422827 CACUGCCCAGCCAAUUUCUGGGUUAAUUUCGUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCAACACGACCACCACCA------ ((.((((((((..((((((.((((((((((((.....))))..)))))))).))))))...))))...)))).))...................------ ( -20.60, z-score = -1.74, R) >droAna3.scaffold_13117 2809449 79 - 5790199 CACUGCCCAGCCGAUUUCUGGGUUAAUUUCAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCG--------------------- ((.((((((((..((((((.((((((((((((.....))))..)))))))).))))))...))))...)))).))....--------------------- ( -22.10, z-score = -3.61, R) >droEre2.scaffold_4690 15943393 100 - 18748788 CACUGCCCAGCCAAUUUCUGGGUUAAUUUCGUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCACCACCAGCAGCAGCAGCACCA ..((((...(((.((((((.((((((((((((.....))))..)))))))).))))))..((....)))))((((........)))).))))........ ( -27.80, z-score = -2.48, R) >droYak2.chrX 7447371 94 + 21770863 CACUGCCCAGCCAAUUUCUGGGUUAAUUUCGUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCAACAGCACCACCACCA------ ....(((((((..((((((.((((((((((((.....))))..)))))))).))))))...))))...)))((((.....))))..........------ ( -24.50, z-score = -2.76, R) >droSec1.super_24 252019 94 - 912625 CACUGCCCAGCCAAUUUCUGGGUUAAUUUCGUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCAACACCACCACCAGCA------ ....(((((((..((((((.((((((((((((.....))))..)))))))).))))))...))))...)))((((..............)))).------ ( -23.64, z-score = -2.46, R) >droSim1.chrX 5577969 94 - 17042790 CACUGCCCAGCCAAUUUCUGGGUUAAUUUCGUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCAACACCACCACCAGCA------ ....(((((((..((((((.((((((((((((.....))))..)))))))).))))))...))))...)))((((..............)))).------ ( -23.64, z-score = -2.46, R) >droPer1.super_14 397767 82 - 2168203 CACUGCCCAGGCGAUUUCUGGGUUAAUUUUAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAAGCGCUGACCACCA------------------ .......(((((.((((((.((((((((((...........)))))))))).))))))..((....)).)).))).......------------------ ( -15.90, z-score = -1.26, R) >dp4.chrXL_group1e 10961948 82 - 12523060 CACUGCCCAGGCGAUUUCUGGGUUAAUUUUAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAAGCGCUGACCACCA------------------ .......(((((.((((((.((((((((((...........)))))))))).))))))..((....)).)).))).......------------------ ( -15.90, z-score = -1.26, R) >droVir3.scaffold_12932 956410 97 - 2102469 CACUGCACACAAAAUUUCUGGGUUAAUUUUAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAAGCGCUGACCACCAACGG-GCACAGAGCCA-- ...((((.(((..((((((.((((((((((...........)))))))))).))))))..))))).)).((.(((.((......))-...))).))..-- ( -19.80, z-score = -1.57, R) >droMoj3.scaffold_6473 14771627 98 - 16943266 CACUGCACACCAAAUUUCUGGGUUAAUUUUAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAAGCGCUGACCACCACCGGAGCAGCAGAUCA-- ..((((.((.((.((((((.((((((((((...........)))))))))).))))))..)).))....((.(((........))).)).))))....-- ( -22.60, z-score = -2.39, R) >droGri2.scaffold_14853 5384753 89 + 10151454 CACUGCGCACAAAAUUUCUGGGUUAAUUUUAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAAGCGCUGACCACCACCCGAGC----------- ....((((.....((((((.((((((((((...........)))))))))).))))))..((....)).))))................----------- ( -17.70, z-score = -1.95, R) >consensus CACUGCCCAGCCAAUUUCUGGGUUAAUUUCAUGAAAAAUGAAAAAUUAAUUGAGAAAUAAUGUUGACAGGCGCUGACCACCACCACCACCA_CA______ ((.(((.......((((((.((((((((((((.....))))..)))))))).))))))..((....)).))).))......................... (-14.08 = -13.58 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:46 2011