
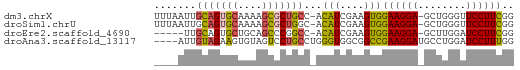
| Sequence ID | dm3.chrX |
|---|---|
| Location | 7,028,572 – 7,028,630 |
| Length | 58 |
| Max. P | 0.940184 |

| Location | 7,028,572 – 7,028,630 |
|---|---|
| Length | 58 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 71.14 |
| Shannon entropy | 0.44543 |
| G+C content | 0.55242 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
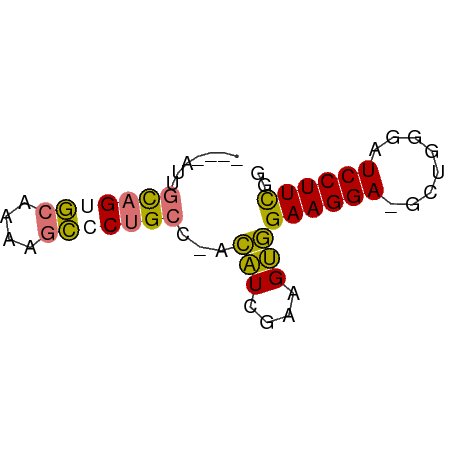
>dm3.chrX 7028572 58 + 22422827 UUUAAUUGCAGUGCAAAAGCGCUGCC-ACAUCGAAGUGGAAGGA-GCUGGGUUCCUUCGG .......(((((((....)))))))(-((......)))((((((-((...)))))))).. ( -23.80, z-score = -3.30, R) >droSim1.chrU 9966838 58 + 15797150 UUUAAUUGCAGUGCAAAAGCGCUGGC-ACAUCGAAGUGGAAGGA-GCUGGGUUCCUUCGG ........((((((....)))))).(-((......)))((((((-((...)))))))).. ( -21.00, z-score = -2.46, R) >droEre2.scaffold_4690 15929864 53 + 18748788 -----UUGCAGUGCUGCAGCCCGGCC-ACAUCGAAGUGGAAGGA-GCUUGGAUCCUUCGG -----.((((....))))((((..((-((......))))..)).-))............. ( -15.50, z-score = 0.30, R) >droAna3.scaffold_13117 2797739 56 + 5790199 ----AUUGUAGAAGUGUAGUCCUGCCUGGGUGGCGGCCGAAGGAUGCCUGGAUCCUUUGG ----.................(((((.....)))))(((((((((......))))))))) ( -18.60, z-score = -1.11, R) >consensus ____AUUGCAGUGCAAAAGCCCUGCC_ACAUCGAAGUGGAAGGA_GCUGGGAUCCUUCGG .......(((((((....)))))))...(((....)))((((((........)))))).. (-13.18 = -12.93 + -0.25)
| Location | 7,028,572 – 7,028,630 |
|---|---|
| Length | 58 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 71.14 |
| Shannon entropy | 0.44543 |
| G+C content | 0.55242 |
| Mean single sequence MFE | -15.12 |
| Consensus MFE | -8.48 |
| Energy contribution | -9.35 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 7028572 58 - 22422827 CCGAAGGAACCCAGC-UCCUUCCACUUCGAUGU-GGCAGCGCUUUUGCACUGCAAUUAAA ..((((((.......-))))))(((......))-)((((.((....)).))))....... ( -18.80, z-score = -2.84, R) >droSim1.chrU 9966838 58 - 15797150 CCGAAGGAACCCAGC-UCCUUCCACUUCGAUGU-GCCAGCGCUUUUGCACUGCAAUUAAA ..((((((.......-))))))(((......))-).(((.((....)).)))........ ( -13.50, z-score = -1.49, R) >droEre2.scaffold_4690 15929864 53 - 18748788 CCGAAGGAUCCAAGC-UCCUUCCACUUCGAUGU-GGCCGGGCUGCAGCACUGCAA----- ((((((((.......-)))))((((......))-)).)))..((((....)))).----- ( -15.20, z-score = -0.06, R) >droAna3.scaffold_13117 2797739 56 - 5790199 CCAAAGGAUCCAGGCAUCCUUCGGCCGCCACCCAGGCAGGACUACACUUCUACAAU---- .....((.(((.(((........)))(((.....))).))))).............---- ( -13.00, z-score = -0.95, R) >consensus CCGAAGGAACCAAGC_UCCUUCCACUUCGAUGU_GGCAGCGCUUCAGCACUGCAAU____ ..((((((........)))))).............((((.((....)).))))....... ( -8.48 = -9.35 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:44 2011