
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,980,362 – 6,980,458 |
| Length | 96 |
| Max. P | 0.720191 |

| Location | 6,980,362 – 6,980,458 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.63681 |
| G+C content | 0.34361 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -9.90 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6980362 96 + 22422827 GUUAGUUCUUUGAAUUUCA--UUGGUUAGA-UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA--AUUGGAUUUGAUUUUUGAU---------UUAA-GCAUUA .......((((((((((..--......)))-)))))))(((..((((.....))))..).......((((--((..((.......))..))---------))))-)).... ( -21.30, z-score = -1.11, R) >droPer1.super_25 517233 99 - 1448063 ----GCCAUUUUUGUUUCAUUUUUUUGGUU-UUUUAAGGCGUGCUUGAAAUUCGAGUGCUGCAAAUAUCGUUAUCAAAGGUAUUUAUUGCU-----CUGGAAGAAGCAU-- ----(((.((...(..(((......)))..-)...)))))((((((....(((.((.((...(((((((.........)))))))...)).-----)).))).))))))-- ( -21.40, z-score = -0.65, R) >dp4.chrXL_group1a 6392121 100 + 9151740 ----GCCAUUUUUGUUUCAUUUUUUUUUUUGUUUUAAGGCGUGCUUGAAAUUCGAGUGCAGCAAAUAUCGUUAUCAAAGGUAUUUAUUGCU-----CUGGAAGAAGCAU-- ----........((((((.............(((((((.....)))))))(((.((.((((.(((((((.........))))))).)))).-----)).))))))))).-- ( -23.90, z-score = -1.79, R) >droAna3.scaffold_13117 2326852 77 + 5790199 -------------------GAACCGUCAGG-CGGCAAGGCGUGCUUGAAACUCGAGUGCAGCAAUCGU-A--CUUGUAUCUUAUC-UUGAU----------UUAAGCCUUA -------------------...(((.....-))).((((((..((((.....))))..).((((....-.--.))))........-.....----------....))))). ( -19.80, z-score = -0.51, R) >droEre2.scaffold_4690 15890842 97 + 18748788 GUUACUUCAUUGAAUUUCA--UUGGUUAGA-UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA--AUUGUAUUUGAUUUUUGAU---------UUAAAGCAUUA (((.....((..((..(((--.......((-((((((((((..((((.....))))..).))..))))))--))).....)))..))..))---------....))).... ( -18.50, z-score = -0.26, R) >droYak2.chrX 7396541 103 - 21770863 GUUAAUUCUUUGAAUUUCA--UUGGUUAGA-UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA--AUUGUAUUUGAUUGUUGAUAUUUGAUUU---AAGCAUUA ......(((((((((((..--......)))-)))))))).(((((((((..(((((((((((((((....--.........)))))))).))))))))))---)))))).. ( -26.22, z-score = -2.67, R) >droSec1.super_24 200536 107 + 912625 GUUAGUUCUUUGAAUUUCACAUUGGUAAGA-UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA--AUAGUAUUUGAUUUUUGAU-UUUGAUUUUUGAAGCAUUA ..((((.(((..((..(((....((((..(-((((((((((..((((.....))))..).))..))))))--))..))))...........-..)))..))..))).)))) ( -19.73, z-score = 0.18, R) >droSim1.chrX 5493374 98 + 17042790 GUUAGUUCUUUGAAUUUCACAUUGGUUAGA-UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA--AUUGUAUUUGAUUUUUGAU---------UUAA-GCAUUA ....((.((((((((((.((....)).)))-)))))))))(((((((((.(((((((((((.........--.)))))))))).....).)---------))))-)))).. ( -22.50, z-score = -1.51, R) >consensus GUUAGUUCUUUGAAUUUCA__UUGGUUAGA_UUUAAGGGCGUGCUUGAAACUCGAGUGCAGCGAUUUUGA__AUUGUAUUUGAUUUUUGAU_________UUAAAGCAUUA ......................................(((..((((.....))))..).))................................................. ( -9.90 = -9.90 + 0.00)

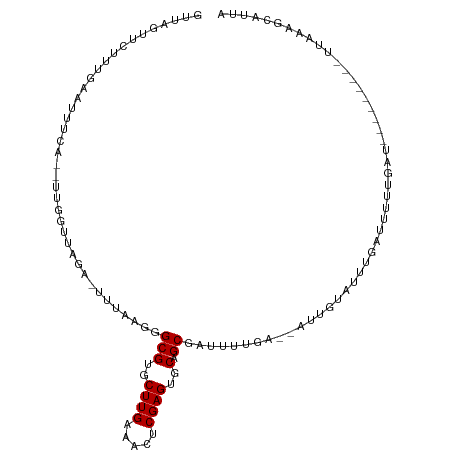
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:40 2011