| Sequence ID | dm3.chr2L |
|---|---|
| Location | 906,015 – 906,107 |
| Length | 92 |
| Max. P | 0.999307 |

| Location | 906,015 – 906,107 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.50095 |
| G+C content | 0.41343 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -13.09 |
| Energy contribution | -12.72 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
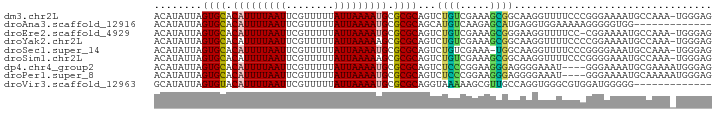
>dm3.chr2L 906015 92 + 23011544 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUGUCGAAAGCGGCAAGGUUUUCCCGGGAAAAUGCCAAA-UGGGAG ........((((.(((((((((........))))))))).))))....(((((....)))))......((((.((......))...-.)))). ( -25.10, z-score = -1.77, R) >droAna3.scaffold_12916 2627508 80 + 16180835 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGCAUGUCAAGAGCAUGAGGUGGAAAAAGGGGGUGG------------- .(((....((((.(((((((((........))))))))).))))..(((.(((......))).)))..........))).------------- ( -20.10, z-score = -2.37, R) >droEre2.scaffold_4929 968750 91 + 26641161 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUGUCGAAAGCGGGAAGGUUUUCC-CGGAAAAUGCCAAA-UGGGAG (((.(((.((((.(((((((((........))))))))).))))))).)))(....)((((((....))))-)).....(.((...-.)).). ( -23.80, z-score = -1.54, R) >droYak2.chr2L 892724 92 + 22324452 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAAGCGCGCAGUCUGUCGAAAGCGGCAAGGUUUUCCCCGGAAAAUGCCAAA-UGGGAG .(((....((((...(((((((........))))))).))))(((.((((..((((((......))))))..))))...)))...)-)).... ( -21.70, z-score = -0.87, R) >droSec1.super_14 877552 91 + 2068291 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUGUCGAAA-UGGCAAGGUUUUCCCGGGGAAAUGCCAAA-UGGGAG ........((((.(((((((((........))))))))).))))....((((....-.))))......(((((.((.....))...-))))). ( -24.70, z-score = -1.74, R) >droSim1.chr2L 895185 92 + 22036055 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAAGCGCGCAGUCUGUCGAAAGCGGCAAGGUUUUCCCGGGGAAAUGCCAAA-UGGGAG ........((((.(.(((((((........))))))).).))))....(((((....)))))......(((((.((.....))...-))))). ( -23.00, z-score = -1.03, R) >dp4.chr4_group2 801172 89 - 1235136 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUCCCGGAAGGGAGGGGAAAU----GGGAAAAUGCGAAAAUGGGAG .(((....((((.(((((((((........))))))))).))))..((((((....))))))....))----).................... ( -27.60, z-score = -3.64, R) >droPer1.super_8 3454957 89 + 3966273 ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUCCCGGAAGGGAGGGGAAAU----GGGAAAAUGCAAAAAUGGGAG .(((....((((.(((((((((........))))))))).))))..((((((....))))))....))----).................... ( -27.60, z-score = -3.88, R) >droVir3.scaffold_12963 19466805 80 + 20206255 GCAUAUUAGUGUACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGGUAAAAAGCGUUGCCAGGUGGGCGUGGAUGGGGG------------- ((.((((.(((..(((((((((........)))))))))..))).(((((......))))).)))).))...........------------- ( -19.40, z-score = -0.81, R) >consensus ACAUAUUAGUGCACAUUUUAAUUCGUUUUUAUUAAAAUGCGCGCAGUCUGUCGAAAGCGGCAAGGUUUUCCCGGGAAAAUGCCAAA_UGGGAG ........((((.(((((((((........))))))))).))))...((((.....))))................................. (-13.09 = -12.72 + -0.37)
| Location | 906,015 – 906,107 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.50095 |
| G+C content | 0.41343 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.16 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
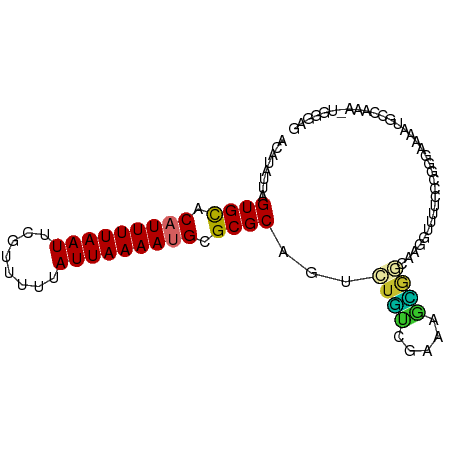
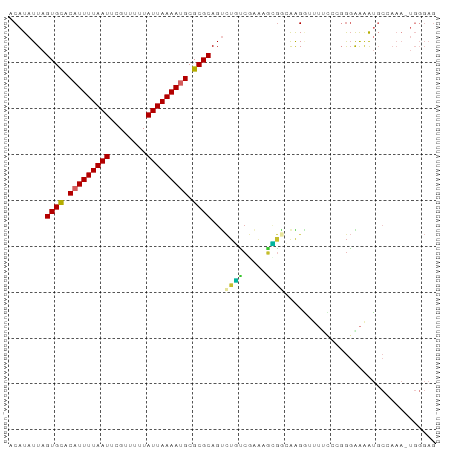
>dm3.chr2L 906015 92 - 23011544 CUCCCA-UUUGGCAUUUUCCCGGGAAAACCUUGCCGCUUUCGACAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ......-...(((((((((....)))))...))))((..((....))..))((((((((((((........)))))))))))).......... ( -25.60, z-score = -3.42, R) >droAna3.scaffold_12916 2627508 80 - 16180835 -------------CCACCCCCUUUUUCCACCUCAUGCUCUUGACAUGCUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU -------------.............................(((((..(.((((((((((((........)))))))))))).)...))))) ( -15.80, z-score = -3.75, R) >droEre2.scaffold_4929 968750 91 - 26641161 CUCCCA-UUUGGCAUUUUCCG-GGAAAACCUUCCCGCUUUCGACAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ....((-(((((........(-((((....)))))((..((....))..))((((((((((((........)))))))))))).)))).))). ( -24.80, z-score = -3.65, R) >droYak2.chr2L 892724 92 - 22324452 CUCCCA-UUUGGCAUUUUCCGGGGAAAACCUUGCCGCUUUCGACAGACUGCGCGCUUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ......-...(((((((((....)))))...))))((..((....))..))((((.(((((((........))))))).)))).......... ( -21.30, z-score = -1.51, R) >droSec1.super_14 877552 91 - 2068291 CUCCCA-UUUGGCAUUUCCCCGGGAAAACCUUGCCA-UUUCGACAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ......-..((((((((((...)))))....)))))-.....(((....(.((((((((((((........)))))))))))).).....))) ( -22.60, z-score = -2.78, R) >droSim1.chr2L 895185 92 - 22036055 CUCCCA-UUUGGCAUUUCCCCGGGAAAACCUUGCCGCUUUCGACAGACUGCGCGCUUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ......-...(((((((((...)))))....))))((..((....))..))((((.(((((((........))))))).)))).......... ( -20.30, z-score = -1.67, R) >dp4.chr4_group2 801172 89 + 1235136 CUCCCAUUUUCGCAUUUUCCC----AUUUCCCCUCCCUUCCGGGAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ...........(((.((((((----................)))))).)))((((((((((((........)))))))))))).......... ( -24.59, z-score = -5.38, R) >droPer1.super_8 3454957 89 - 3966273 CUCCCAUUUUUGCAUUUUCCC----AUUUCCCCUCCCUUCCGGGAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ...........(((.((((((----................)))))).)))((((((((((((........)))))))))))).......... ( -24.29, z-score = -5.21, R) >droVir3.scaffold_12963 19466805 80 - 20206255 -------------CCCCCAUCCACGCCCACCUGGCAACGCUUUUUACCUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUACACUAAUAUGC -------------...........(((.....)))..(((.........))).((((((((((........))))))))))............ ( -13.30, z-score = -2.02, R) >consensus CUCCCA_UUUGGCAUUUUCCCGGGAAAACCUUGCCGCUUUCGACAGACUGCGCGCAUUUUAAUAAAAACGAAUUAAAAUGUGCACUAAUAUGU ...................................................((((((((((((........)))))))))))).......... (-11.82 = -12.16 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:20 2011