| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,941,126 – 6,941,231 |
| Length | 105 |
| Max. P | 0.749283 |

| Location | 6,941,126 – 6,941,231 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Shannon entropy | 0.38846 |
| G+C content | 0.38882 |
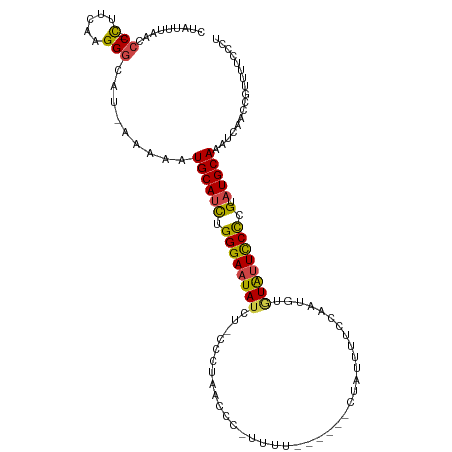
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749283 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
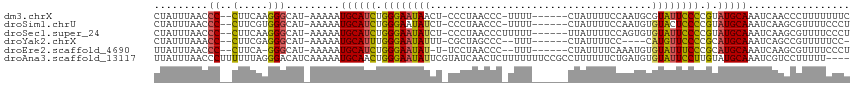
>dm3.chrX 6941126 105 - 22422827 CUAUUUAACCC--CUUCAAGGGCAU-AAAAAUGCAUCUGGGAAUAACU-CCCUAACCC-UUUU------CUAUUUUCCAAUGCGUAUUCCCCGUAUGCAAAUCAACCCUUUUUUUC ...........--....((((((((-....))))....((((.....)-)))....))-))..------...........(((((((.....)))))))................. ( -17.60, z-score = -2.16, R) >droSim1.chrU 674838 105 + 15797150 CUAUUUAACCC--CUUCGUGGGCAU-AAAAAUGCAUCUGGGAAUAUCU-CCCUAACCC-UUUU------CUAUUUUCCAAUGUGUACUCCCCGUAUGCAAAUCAAGCGUUUUCCCU ...........--......(((...-.(((((((....((((.....)-)))......-....------...........(((((((.....)))))))......)))))))))). ( -20.60, z-score = -2.73, R) >droSec1.super_24 161272 106 - 912625 CUAUUUAACCC--CUUCAAGGGCAU-AAAAAUGCAUCUGGGAAUAUCU-CCCUAACCCUUUUU------UUAUUUUCCAGUGUGUAUUCCCCGUAUGCAAAUCAAGCGUUUUCCCU ...........--.....((((...-.(((((((..(((((((.....-..............------....)))))))(((((((.....)))))))......))))))))))) ( -19.60, z-score = -1.82, R) >droYak2.chrX 7354313 99 + 21770863 CUAUUUAAACC--CUUCGAGGGCAU-AAAAAUGCAUUUGGGAAUAUUU-CGCUAGCCC--UUU------CUAUUUUCC----CAUGUUCCCCGCAUGCAAAUCAGCCGUUUUUCC- ...((((..((--(.....)))..)-)))..(((((..((((((((..-...(((...--...------)))......----.))))))))...)))))................- ( -17.60, z-score = -0.32, R) >droEre2.scaffold_4690 15852535 102 - 18748788 UUAUUUAACCC--CUUCA-GGGCAU-AAAAAUGCAUCUGGGAAUAU-U-UCCUAACCC--UUU------CUAUUUUCAAAUGUGUAUUUCCCGCAUGCAAAUCAAGCGUUUUCCCU ...........--....(-(((...-.(((((((...(((((....-.-)))))....--...------..........(((((.......))))).........))))))))))) ( -17.20, z-score = -0.96, R) >droAna3.scaffold_13117 3505222 112 + 5790199 UUAUUUAACCCUUUUUUAGGGACAUCAAAAAUGCAACUGGGAAUAUUCGUAUCAACUCUUUUUUUCCGCCUUUUUUCUGAUGUGUAUUCCUUGUAUGCAAAUCGUCCUUUUU---- ........((((.....))))..........((((((.((((((((...(((((.......................))))).)))))))).)).)))).............---- ( -20.70, z-score = -2.09, R) >consensus CUAUUUAACCC__CUUCAAGGGCAU_AAAAAUGCAUCUGGGAAUAUCU_CCCUAACCC_UUUU______CUAUUUUCCAAUGUGUAUUCCCCGUAUGCAAAUCAACCGUUUUCCCU ........(((........))).........((((((.((((((((.....................................)))))))).).)))))................. (-11.60 = -11.77 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:35 2011