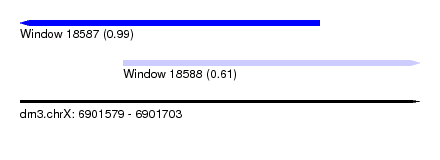
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,901,579 – 6,901,703 |
| Length | 124 |
| Max. P | 0.990095 |

| Location | 6,901,579 – 6,901,672 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.34 |
| Shannon entropy | 0.60881 |
| G+C content | 0.38681 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -10.02 |
| Energy contribution | -8.89 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.75 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
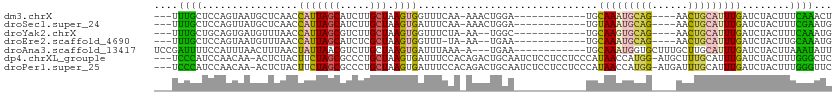
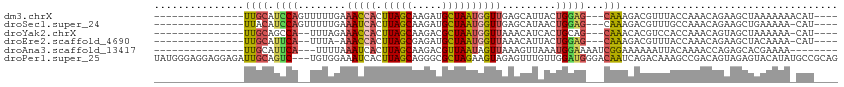
>dm3.chrX 6901579 93 - 22422827 ---UUUGCUCCAGUAAUGCUCAACCAUUAGCAUCUUGCUAAGUGGUUUCAA-AAACUGGA------------UGCAAAUGCAG----AACUGCAUUUGAUCUACUUUCAAACU ---(((((((((((.......((((((((((.....))).)))))))....-..))))))------------.)))))((((.----...))))(((((.......))))).. ( -28.22, z-score = -3.91, R) >droSec1.super_24 128814 93 - 912625 ---UUUGCUCCAGUUAUGCUCAACCAUUAGCAUCUUGCUAAGUGAUUUCAA-AAACUGGA------------UGUAAAUGCAG----AACUGCAUUUGAUCUACUUUCGAAUG ---((((((((((((.((.(((....(((((.....))))).)))...)).-.)))))))------------.))))).(((.----...)))((((((.......)))))). ( -23.00, z-score = -2.54, R) >droYak2.chrX 7322426 91 + 21770863 ---UUUGCUGCAGUGAUGUUUAACCAUUAGCGUCUUGCUAAGUGGUUUCUA-AA--UGGC------------UGCAAGUGCAG----AACUGCAUUUGAUCUACUUUCAAAUG ---(((((((((((.((....((((((((((.....))).)))))))....-.)--).))------------))))...))))----)....(((((((.......))))))) ( -29.10, z-score = -2.93, R) >droEre2.scaffold_4690 15823663 90 - 18748788 ---UUUGCUCCAGUAAUGUUUAACCAUUAGCAUCUCGCUAAGUGGUUU-UA-AA--UGAA------------UGCAAAUGCAG----AACUGCAUUUGAUCUACUUGCAAAUG ---(((((...((((..((((((((((((((.....))).))))))..-))-))--)...------------..((((((((.----...))))))))...)))).))))).. ( -25.20, z-score = -3.18, R) >droAna3.scaffold_13417 387252 97 - 6960332 UCCGAUUUUCCAUUUAACUUUAACUAUUAACGUCUUGCUAAGUGAUUUAAA-A---UGAA------------UGCAAAUGGUGCUUUGCUUGCAUUUGAUCUACUUAAAUAUU ......................................(((((((((..((-(---((..------------.(((((......)))))...))))))))).)))))...... ( -13.20, z-score = -0.55, R) >dp4.chrXL_group1e 6791213 108 - 12523060 ---UCCCAUCCAACAA-ACUCUACUUCUAGCGCCCUGCUAAGUGAUUUCCACAGACUGCAAUCUCCUCCUCCCAUAACCAUGG-AUGCUUUGCAUUUGAUCUACUUUGGGCUC ---.............-..............((((....(((((((.....((((.(((((...(.(((............))-).)..)))))))))))).)))).)))).. ( -18.80, z-score = -1.02, R) >droPer1.super_25 126543 108 - 1448063 ---UCCCAUCCAACAA-ACUCUACUUCUAGCGCCCUGCUAAGUGAUUUCCACAGACUGCAAUCUCCUCCUCCCAUAACCAUGG-AUGAUUUGCAUUUGAUCUACUUUGGGUUC ---.((((........-....((((..((((.....)))))))).......((((.(((((..((.(((............))-).)).)))))))))........))))... ( -19.80, z-score = -1.24, R) >consensus ___UUUGCUCCAGUAAUGCUCAACCAUUAGCGUCUUGCUAAGUGAUUUCAA_AAACUGGA____________UGCAAAUGCAG____AACUGCAUUUGAUCUACUUUCAAAUG ....((((................(((((((.....))).))))..............................(((((((((......)))))))))........))))... (-10.02 = -8.89 + -1.14)
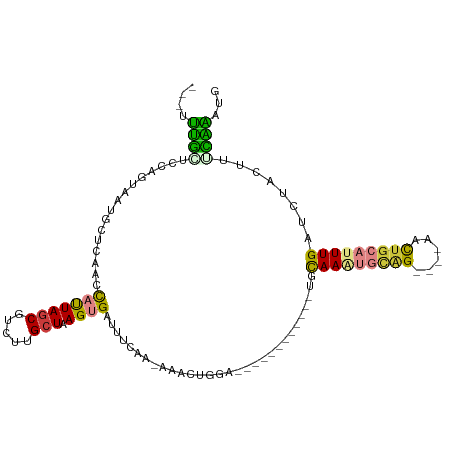
| Location | 6,901,611 – 6,901,703 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.68 |
| Shannon entropy | 0.61778 |
| G+C content | 0.37842 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -7.49 |
| Energy contribution | -7.63 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6901611 92 + 22422827 ---------------UUGCAUCCAGUUUUUGAAACCACUUAGCAAGAUGCUAAUGGUUGAGCAUUACUGGAG---CAAAGACGUUUACCAAACAGAAGCUAAAAAAACAU---- ---------------((((.((((((...((.(((((.(((((.....))))))))))...))..)))))))---)))................................---- ( -25.40, z-score = -3.72, R) >droSec1.super_24 128846 91 + 912625 ---------------UUACAUCCAGUUUUUGAAAUCACUUAGCAAGAUGCUAAUGGUUGAGCAUAACUGGAG---CAAAGACGUUUGCCAAACAGAAGCUGAAAAA-CAU---- ---------------.....(((((((..((.(((((.(((((.....))))))))))...)).)))))))(---((((....)))))....(((...))).....-...---- ( -21.90, z-score = -2.11, R) >droYak2.chrX 7322458 89 - 21770863 ---------------UUGCAGCCA--UUUAGAAACCACUUAGCAAGACGCUAAUGGUUAAACAUCACUGCAG---CAAACACGUCCACCAAACAGUAGCUAAAAAA-CAU---- ---------------((((((...--......(((((.(((((.....))))))))))........))))))---...............................-...---- ( -15.43, z-score = -1.50, R) >droEre2.scaffold_4690 15823695 88 + 18748788 ---------------UUGCAUUCA--UUUA-AAACCACUUAGCGAGAUGCUAAUGGUUAAACAUUACUGGAG---CAAAGACGUUUACCAAACAGAAGCUACAAAA-CAU---- ---------------..((.(((.--....-.(((((.(((((.....)))))))))).........(((((---(......)))..)))....))))).......-...---- ( -14.70, z-score = -1.49, R) >droAna3.scaffold_13417 387288 88 + 6960332 ---------------UUGCAUUCA---UUUUAAAUCACUUAGCAAGACGUUAAUAGUUAAAGUUAAAUGGAAAAUCGGAAAAAAUUACAAAACCAGAGCACGAAAA-------- ---------------.(((.((((---(((......(((((((.....))))..))).......))))))).....((..............))...)))......-------- ( -7.76, z-score = 0.50, R) >droPer1.super_25 126579 111 + 1448063 UAUGGGAGGAGGAGAUUGCAGUC---UGUGGAAAUCACUUAGCAGGGCGCUAGAAGUAGAGUUUGUUGGAUGGGACAAUCAGACAAAGCCGACAGUAGAGUACAUAUGCCGCAG ..((.((........)).))..(---(((((.....((((.((..(((.(((....)))..((((((.(((......))).)))))))))....)).)))).......)))))) ( -21.70, z-score = 1.11, R) >consensus _______________UUGCAUCCA__UUUUGAAACCACUUAGCAAGACGCUAAUGGUUAAACAUAACUGGAG___CAAAGACGUUUACCAAACAGAAGCUAAAAAA_CAU____ ....................((((........(((((.(((((.....)))))))))).........))))........................................... ( -7.49 = -7.63 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:32 2011