| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,884,847 – 6,884,940 |
| Length | 93 |
| Max. P | 0.862596 |

| Location | 6,884,847 – 6,884,940 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 63.39 |
| Shannon entropy | 0.69264 |
| G+C content | 0.49838 |
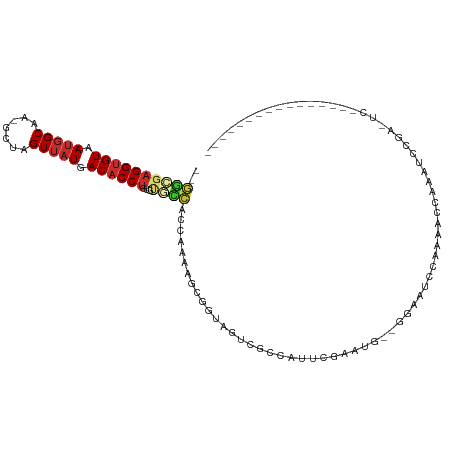
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.94 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
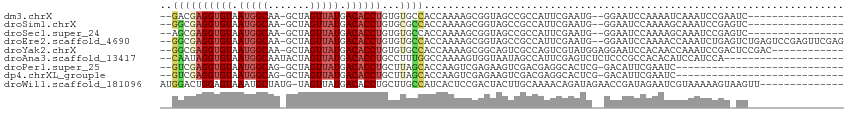
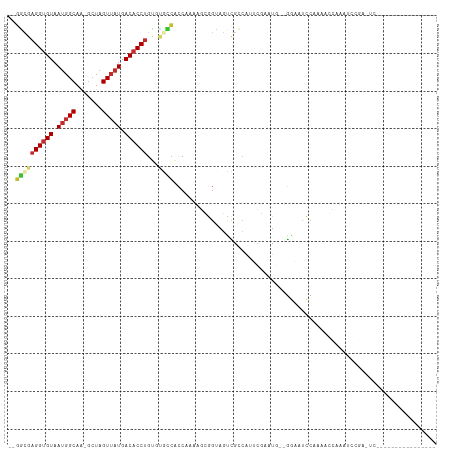
>dm3.chrX 6884847 93 - 22422827 --GACGAGGUGUAAUGGCAA-GCUAGUUAUGACACCUGUGUGCCACCAAAAGCGGUAGCCGCCAUUCGAAUG--GGAAUCCAAAAUCAAAUCCGAAUC---------------- --....((((((.(((((..-....))))).))))))(((.((.(((......))).))))).(((((.((.--.((........))..)).))))).---------------- ( -23.90, z-score = -1.04, R) >droSim1.chrX 5407129 93 - 17042790 --GGCGAGGUGUAAUGGCAA-GCUAGUUAUGACACCUGUGCGCCACCAAAAGCGGUAGCCGCCAUUCGAAUG--GGAAUCCAAAAGCAAAUCCGAGUC---------------- --((((((((((.(((((..-....))))).))))))....((.(((......))).)))))).......((--(....)))................---------------- ( -26.20, z-score = -0.57, R) >droSec1.super_24 112160 93 - 912625 --AGCGAGGUGUAAUGGCAA-GCUAGUUAUGACACCUGUGUGCCACCAAAAGCGGUAGCCGCCAUUCGAAUG--GGAAUCCAAAAGCAAAUCCGAGUC---------------- --.((.((((((.(((((..-....))))).))))))(((.((.(((......))).)))))........((--(....)))...))...........---------------- ( -23.90, z-score = -0.41, R) >droEre2.scaffold_4690 15806746 109 - 18748788 --GGCGAGGUGUAAUGGCAA-GCUAGUUAUGACACCUGUGUGCCACCAAAAGCGGUAGCCGCCAUUCGAAUG--GGAAUCCAAAACCAAAUCUGAGUCUGAGUCCGAGUUCGAG --((((((((((.(((((..-....))))).))))))....((.(((......))).))))))..((((((.--(((.((....((((....)).))..)).)))..)))))). ( -34.10, z-score = -1.84, R) >droYak2.chrX 7304917 99 + 21770863 --GGCGAGGUGUAAUGGCAA-GCUAGUUAUGACACCUGUGUGCCACCAAAAGCGGCAGUCGCCAGUCGUAUGGAGGAAUCCACAACCAAAUCCGACUCCGAC------------ --((((((((((.(((((..-....))))).)))))....((((.(.....).)))).))))).((((...(..(((.............)))..)..))))------------ ( -31.42, z-score = -1.40, R) >droAna3.scaffold_13417 363956 92 - 6960332 --CAAUAGGUGUAAUGGCAAUACUAGUUAUGACACCUGCCUUUGGCCAAAAGUGGUAAUAGCCAUUCGAGUCUCUCCCGCCACACAUCCAUCCA-------------------- --...(((((((.(((((.......))))).)))))))....((((....((((((....)))))).(((...)))..))))............-------------------- ( -22.30, z-score = -1.32, R) >droPer1.super_25 106295 82 - 1448063 --GUCGAGGUGUAAUGGCAG-GCUAGUUAUGACACCUGCUUAGCACCAAGUCGAGAAGUCGACGAGGCACUCG-GACAUUCGAAUC---------------------------- --.(((((((((...(((((-(............))))))..)))))..((((((..(((.....))).))).-)))..))))...---------------------------- ( -27.90, z-score = -2.13, R) >dp4.chrXL_group1e 6770875 82 - 12523060 --GUCGAGGUGUAAUGGCAG-GCUAGUUAUGACACCUGCUUAGCACCAAGUCGAGAAGUCGACGAGGCACUCG-GACAUUCGAAUC---------------------------- --.(((((((((...(((((-(............))))))..)))))..((((((..(((.....))).))).-)))..))))...---------------------------- ( -27.90, z-score = -2.13, R) >droWil1.scaffold_181096 4790819 99 - 12416693 AUGGACUGGAGUAAAUGCUAUG-UAGUUAUGACACCUGCUUGCCAUCACUCCGACUACUUGCAAAACAGAUAGAACCGAUAGAAUCGUAAAAAGUAAGUU-------------- ......((((((....((...(-(((.........))))..))....))))))...((((((....(.....)...((((...))))......)))))).-------------- ( -15.40, z-score = 0.57, R) >consensus __GGCGAGGUGUAAUGGCAA_GCUAGUUAUGACACCUGUGUGCCACCAAAAGCGGUAGUCGCCAUUCGAAUG__GGAAUCCAAAACCAAAUCCGA_UC________________ ..((((((((((.(((((.......))))).))))))...))))...................................................................... (-11.51 = -11.94 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:30 2011