| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,879,265 – 6,879,329 |
| Length | 64 |
| Max. P | 0.500000 |

| Location | 6,879,265 – 6,879,329 |
|---|---|
| Length | 64 |
| Sequences | 7 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Shannon entropy | 0.49483 |
| G+C content | 0.39907 |
| Mean single sequence MFE | -12.74 |
| Consensus MFE | -7.09 |
| Energy contribution | -8.53 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
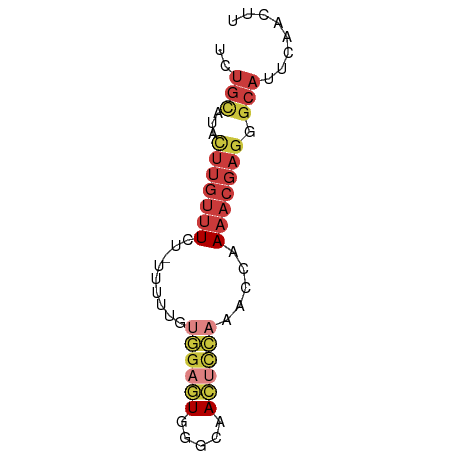
>dm3.chrX 6879265 64 - 22422827 UCUGCAUACUUGUUUCU-UUUUUGUGGAGUGGGCAACUCCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((..-......((((((.....)))))).....))))))).)))........ ( -17.32, z-score = -1.95, R) >droAna3.scaffold_13417 356535 52 - 6960332 -------------UUAUAAACUUUCCUGUUCUUUGAAACCAAACCAAAACGAGGGCAUUCAACUU -------------..........(((((((.((((....))))....))).)))).......... ( -3.60, z-score = 0.60, R) >droYak2.chrX 7299305 64 + 21770863 UCUGCAUACUUGUUUCU-CUUUCGUGGAGUGGGCAACUCCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((..-......((((((.....)))))).....))))))).)))........ ( -17.32, z-score = -2.16, R) >droEre2.scaffold_4690 15801160 64 - 18748788 UUUGCAUACUUGUUUCU-UUUUCGUGGAGUGGGCAACUCCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((..-......((((((.....)))))).....))))))).)))........ ( -17.42, z-score = -2.21, R) >droSec1.super_24 106721 64 - 912625 UCUGUAUACUUGUUUCU-UUUUUGUGGAGUGGGCAACUUCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((..-......((((((.....)))))).....))))))).)))........ ( -12.82, z-score = -0.64, R) >droSim1.chrX 5401632 64 - 17042790 UCUGUAUACUUGUUUCU-UUUUUGUGGAGUGGGCAACUUCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((..-......((((((.....)))))).....))))))).)))........ ( -12.82, z-score = -0.64, R) >droWil1.scaffold_181096 4781204 65 - 12416693 UAUGUAUGUUUGUAUCUACCUUUACUGACUUACCAACACACAUACCAAACGAGGGCAUUCAACUU (((((.((((.(((((..........))..))).)))).)))))((......))........... ( -7.90, z-score = -0.31, R) >consensus UCUGCAUACUUGUUUCU_UUUUUGUGGAGUGGGCAACUCCAAACCAAAACGAGGGCAUUCAACUU ..(((...(((((((.........((((((.....)))))).....))))))).)))........ ( -7.09 = -8.53 + 1.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:28 2011