| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,844,012 – 6,844,108 |
| Length | 96 |
| Max. P | 0.883116 |

| Location | 6,844,012 – 6,844,108 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.70 |
| Shannon entropy | 0.62235 |
| G+C content | 0.44950 |
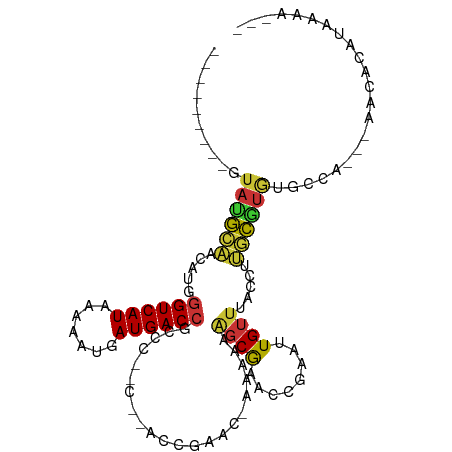
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.20 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
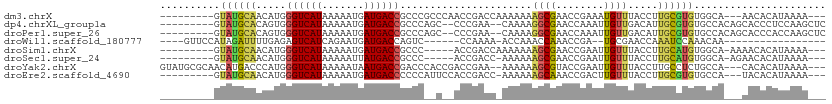
>dm3.chrX 6844012 96 - 22422827 ---------GUAUGCAACAUGGGUCAUAAAAAUGAUGACCGCCCGCCCAACCGACCAAAAAAAGCGAACCGAAAUGUUUACCUUGCGUGUGGCA---AACACAUAAAA--- ---------((.(((.(((((((((((.......))))))...(((.................)))...................))))).)))---.))........--- ( -19.23, z-score = -0.51, R) >dp4.chrXL_group1a 797190 98 - 9151740 ---------GUAUGCACAGUGGGUCAUAAAAAUGAUGACCGCCCAGC--CCCGAA--CAAAAGGCGAACCAAAUUGUUGACAUUGCGUGUGCCACAGCACCCUCCAAGCUC ---------((((((.(((((((((((.......)))))).....((--(.....--.....)))...............))))).))))))...(((.........))). ( -22.80, z-score = -0.27, R) >droPer1.super_26 910276 98 - 1349181 ---------GUAUGCACAGUGGGUCAUAAAAAUGAUGACCGCCCAGC--CCCGAA--CAAAAGGCGAACCAAAUUGUUGACAUUGCGUGUGCCACAGCACCCACCAAGCUC ---------((((((.(((((((((((.......)))))).....((--(.....--.....)))...............))))).))))))...(((.........))). ( -22.80, z-score = -0.22, R) >droWil1.scaffold_180777 2939819 81 - 4753960 ----GUUCCAUAGAUUUUGAGAGUCAUCAGAAUGAUGACCAGUC------CCAAAA-ACCAAACCAAACCGA--UGCGAACCAAAUCCAAACAA----------------- ----(((((((...(((((.((((((((.....))))))...))------.)))))-.......(.....))--)).)))).............----------------- ( -12.50, z-score = -2.53, R) >droSim1.chrX 5373797 93 - 17042790 ---------GUAUGCAACAUGGGUCAUAAAAAUGAUGACCGCCC-----ACCGACCAAAAAAAGCGAACCGAAUUGUUUACCUUGCAUGUGGCA-AAAACACAUAAAA--- ---------...(((.(((((((((((.......))))))....-----............((((((......))))))......))))).)))-.............--- ( -19.00, z-score = -1.18, R) >droSec1.super_24 77564 92 - 912625 ---------GUAUGCAACAUGGGUCAUAAAAAUUAUGACCGCCC-----ACCGACC-AAAAAAGCGAACCGAAUUGUUUACCUUGCAUGUGGCA-AGAACACAUAAAA--- ---------...(((.((((((((((((.....)))))))....-----.......-....((((((......))))))......))))).)))-.............--- ( -20.30, z-score = -1.68, R) >droYak2.chrX 7262049 103 + 21770863 GUAUGCGCAACAUGACCCAUGGGUCAUAAAAAUAAUGACCGACCCACCGACCGAA--AAAAAAGCGUACCGAAUUGUUUACCUUGCCUCUGCCA---CACACAUAAAA--- ((((((....((((...))))((((((.......))))))...............--......)))))).........................---...........--- ( -16.60, z-score = -0.78, R) >droEre2.scaffold_4690 15771796 95 - 18748788 ---------GUAUGCAACAUGGGUCAUAAAAAUGAUGACCCCCCAUUCCACCGACC-AAAAAAGCAAACCGACUUGUUUACCUUGCGUGUGCCA---UACACAUAAAA--- ---------(((((..(((((((((((.......)))))))...............-....((((((......)))))).......))))..))---)))........--- ( -20.70, z-score = -2.38, R) >consensus _________GUAUGCAACAUGGGUCAUAAAAAUGAUGACCGCCC__C__ACCGAAC_AAAAAAGCGAACCGAAUUGUUUACCUUGCGUGUGCCA___AACACAUAAAA___ ..........((((((.....((((((.......))))))......................((((........)))).....))))))...................... ( -9.54 = -9.20 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:25 2011