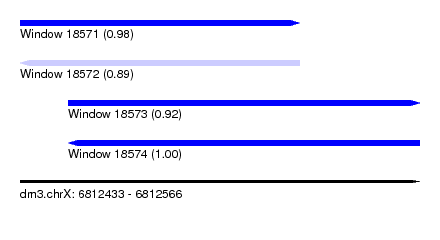
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,812,433 – 6,812,566 |
| Length | 133 |
| Max. P | 0.997863 |

| Location | 6,812,433 – 6,812,526 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.26755 |
| G+C content | 0.56141 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
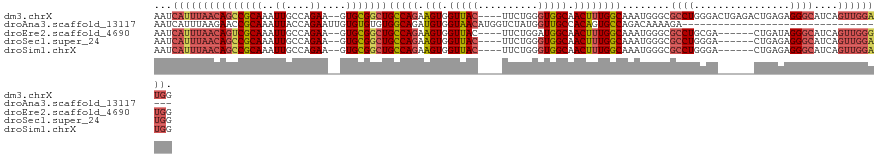
>dm3.chrX 6812433 93 + 22422827 ACCAAGGCAACCGCCGCCAUCCAACUGAUGCCCUCUCAGUCUCAGUCCCAGGCGCCCAUUUGCCAAAGUUGCCACCC-AGAAGUAACCACUUCU .....((((((....(((.....(((((.((.......)).)))))....)))((......))....))))))....-((((((....)))))) ( -24.10, z-score = -3.28, R) >droYak2.chrX 7229238 76 - 21770863 ACCAAGGCAACCAAUGCCAACCAACUGAUACCCCCU-AGUC----------GACCAAAGUUGCCAAAGUUGCCACCCCAGACGUAAC------- .....((((((...((.((((....((((.......-.)))----------)......)))).))..))))))..............------- ( -13.60, z-score = -2.30, R) >droSec1.super_24 26344 87 + 912625 ACCAAGGCAACCGCCGCCAUCCAACUGAUGCCCUCUCAGUC------CCAGGCGCCCAUUUGCCAAAGUUGCCACCC-AGAAGUAACCACUUCU .....((((((....(((.....(((((.......))))).------...)))((......))....))))))....-((((((....)))))) ( -22.60, z-score = -3.16, R) >droSim1.chrX 5342909 87 + 17042790 ACCAAGGCAACCGCCGCCAUCCAACUGAUGCCCUCUCAGUC------CCAGGCGCCCAUUUGCCAAAGUUGCCACCC-AGAAGUAACCACUUCU .....((((((....(((.....(((((.......))))).------...)))((......))....))))))....-((((((....)))))) ( -22.60, z-score = -3.16, R) >consensus ACCAAGGCAACCGCCGCCAUCCAACUGAUGCCCUCUCAGUC______CCAGGCGCCCAUUUGCCAAAGUUGCCACCC_AGAAGUAACCACUUCU .....((((((............(((((.......)))))..........((((......))))...))))))..................... (-13.30 = -14.30 + 1.00)


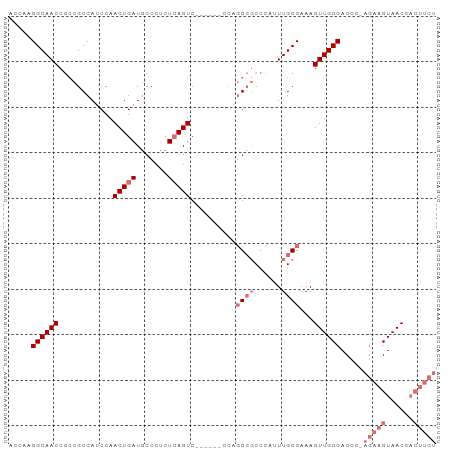
| Location | 6,812,433 – 6,812,526 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.26755 |
| G+C content | 0.56141 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -21.52 |
| Energy contribution | -23.02 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6812433 93 - 22422827 AGAAGUGGUUACUUCU-GGGUGGCAACUUUGGCAAAUGGGCGCCUGGGACUGAGACUGAGAGGGCAUCAGUUGGAUGGCGGCGGUUGCCUUGGU .(((((.((((((...-.)))))).)))))(((((.(.(.((((...((((((..((....))...))))))....)))).).))))))..... ( -36.30, z-score = -2.47, R) >droYak2.chrX 7229238 76 + 21770863 -------GUUACGUCUGGGGUGGCAACUUUGGCAACUUUGGUC----------GACU-AGGGGGUAUCAGUUGGUUGGCAUUGGUUGCCUUGGU -------(((((.......))))).(((..(((((((...(((----------((((-((..(....)..)))))))))...)))))))..))) ( -28.40, z-score = -3.29, R) >droSec1.super_24 26344 87 - 912625 AGAAGUGGUUACUUCU-GGGUGGCAACUUUGGCAAAUGGGCGCCUGG------GACUGAGAGGGCAUCAGUUGGAUGGCGGCGGUUGCCUUGGU .(((((.((((((...-.)))))).)))))(((((.(.(.((((...------((((((.......))))))....)))).).))))))..... ( -32.50, z-score = -1.81, R) >droSim1.chrX 5342909 87 - 17042790 AGAAGUGGUUACUUCU-GGGUGGCAACUUUGGCAAAUGGGCGCCUGG------GACUGAGAGGGCAUCAGUUGGAUGGCGGCGGUUGCCUUGGU .(((((.((((((...-.)))))).)))))(((((.(.(.((((...------((((((.......))))))....)))).).))))))..... ( -32.50, z-score = -1.81, R) >consensus AGAAGUGGUUACUUCU_GGGUGGCAACUUUGGCAAAUGGGCGCCUGG______GACUGAGAGGGCAUCAGUUGGAUGGCGGCGGUUGCCUUGGU .......((((((.....)))))).(((..(((((.(.(.((((.........((((((.......))))))....)))).).))))))..))) (-21.52 = -23.02 + 1.50)

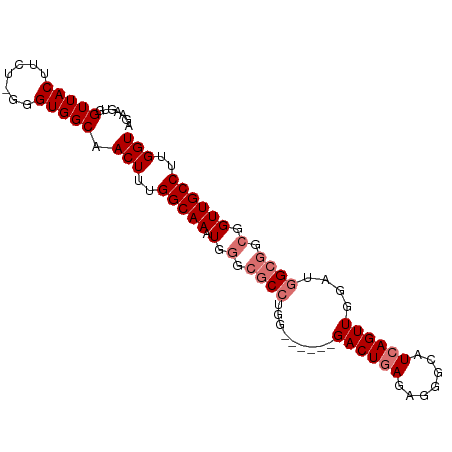

| Location | 6,812,449 – 6,812,566 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Shannon entropy | 0.41494 |
| G+C content | 0.50539 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6812449 117 + 22422827 CCAUCCAACUGAUGCCCUCUCAGUCUCAGUCCCAGGCGCCCAUUUGCCAAAGUUGCCACCCAGAA----GUAACCACUUCUGGCAGCCGCAC--UUCUGGCAAUUUGCGGCUGUUAAAUGAUU .......(((((.((.......)).)))))..(((.(((....((((((((((.((...((((((----((....)))))))).....))))--)).))))))...))).))).......... ( -37.20, z-score = -2.96, R) >droAna3.scaffold_13117 1665492 88 - 5790199 -----------------------------------UCUUUUGUCUGGCACUGUGGCAACCAUAGACCAUGUUACCACAUCUGCCACACACACAAUUCUGGUAAUUUGCGGUUCUUAAAUGAUU -----------------------------------.....(((.(((((.(((((.(((..........))).)))))..))))).))).................................. ( -16.30, z-score = 0.23, R) >droEre2.scaffold_4690 15738130 111 + 18748788 CCACCCAACUGAUGCCCUAUCAG------UCGCAGGCGCCCAUUUGCCAAAGUUGCCAUCCAGAA----GUAACCACUUCUGGCAGCCGCAC--UUCUGGCAAUUUGCGACUGUUAAAUGAUU ....................(((------((((.((((......)))).(((((((((..(((((----((....)))))))((....))..--...)))))))))))))))).......... ( -37.10, z-score = -3.77, R) >droSec1.super_24 26360 111 + 912625 CCAUCCAACUGAUGCCCUCUCAG------UCCCAGGCGCCCAUUUGCCAAAGUUGCCACCCAGAA----GUAACCACUUCUGGCAGCCGCAC--UUCUGGCAAUUUGCGGCUGUUAAAUGAUU .......(((((.......))))------)..(((.(((....((((((((((.((...((((((----((....)))))))).....))))--)).))))))...))).))).......... ( -35.70, z-score = -2.89, R) >droSim1.chrX 5342925 111 + 17042790 CCAUCCAACUGAUGCCCUCUCAG------UCCCAGGCGCCCAUUUGCCAAAGUUGCCACCCAGAA----GUAACCACUUCUGGCAGCCGCAC--UUCUGGCAAUUUGCGGCUGUUAAAUGAUU .......(((((.......))))------)..(((.(((....((((((((((.((...((((((----((....)))))))).....))))--)).))))))...))).))).......... ( -35.70, z-score = -2.89, R) >consensus CCAUCCAACUGAUGCCCUCUCAG______UCCCAGGCGCCCAUUUGCCAAAGUUGCCACCCAGAA____GUAACCACUUCUGGCAGCCGCAC__UUCUGGCAAUUUGCGGCUGUUAAAUGAUU ................................(((.(((.....(((((..(((((.............)))))......)))))((((........)))).....))).))).......... (-15.38 = -15.62 + 0.24)


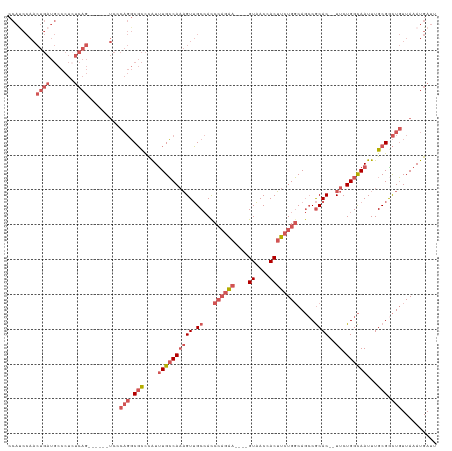
| Location | 6,812,449 – 6,812,566 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Shannon entropy | 0.41494 |
| G+C content | 0.50539 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -25.05 |
| Energy contribution | -26.59 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.997863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
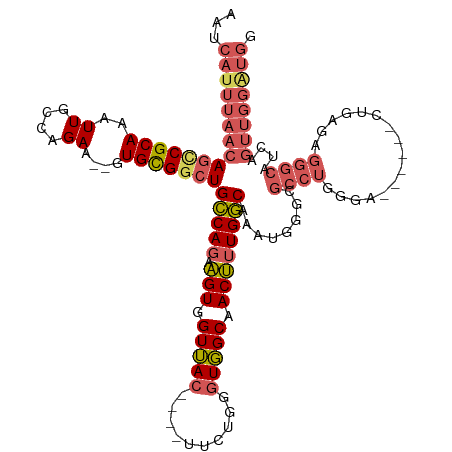
>dm3.chrX 6812449 117 - 22422827 AAUCAUUUAACAGCCGCAAAUUGCCAGAA--GUGCGGCUGCCAGAAGUGGUUAC----UUCUGGGUGGCAACUUUGGCAAAUGGGCGCCUGGGACUGAGACUGAGAGGGCAUCAGUUGGAUGG ...(((((..(((.(((...((((((((.--.(((.((..((((((((....))----)))))))).)))..))))))))....))).))).((((((..((....))...))))))))))). ( -49.20, z-score = -3.58, R) >droAna3.scaffold_13117 1665492 88 + 5790199 AAUCAUUUAAGAACCGCAAAUUACCAGAAUUGUGUGUGUGGCAGAUGUGGUAACAUGGUCUAUGGUUGCCACAGUGCCAGACAAAAGA----------------------------------- ..............(((((.((....)).)))))(((.(((((..(((((((((..........))))))))).))))).))).....----------------------------------- ( -27.30, z-score = -2.35, R) >droEre2.scaffold_4690 15738130 111 - 18748788 AAUCAUUUAACAGUCGCAAAUUGCCAGAA--GUGCGGCUGCCAGAAGUGGUUAC----UUCUGGAUGGCAACUUUGGCAAAUGGGCGCCUGCGA------CUGAUAGGGCAUCAGUUGGGUGG ...((((...(((.(((...((((((((.--((...((((((((((((....))----)))))).)))).))))))))))....))).)))(((------(((((.....)))))))))))). ( -44.40, z-score = -3.07, R) >droSec1.super_24 26360 111 - 912625 AAUCAUUUAACAGCCGCAAAUUGCCAGAA--GUGCGGCUGCCAGAAGUGGUUAC----UUCUGGGUGGCAACUUUGGCAAAUGGGCGCCUGGGA------CUGAGAGGGCAUCAGUUGGAUGG ...((((((((.(((.((..((((((((.--.(((.((..((((((((....))----)))))))).)))..)))))))).)))))((((....------......))))....)))))))). ( -47.40, z-score = -3.50, R) >droSim1.chrX 5342925 111 - 17042790 AAUCAUUUAACAGCCGCAAAUUGCCAGAA--GUGCGGCUGCCAGAAGUGGUUAC----UUCUGGGUGGCAACUUUGGCAAAUGGGCGCCUGGGA------CUGAGAGGGCAUCAGUUGGAUGG ...((((((((.(((.((..((((((((.--.(((.((..((((((((....))----)))))))).)))..)))))))).)))))((((....------......))))....)))))))). ( -47.40, z-score = -3.50, R) >consensus AAUCAUUUAACAGCCGCAAAUUGCCAGAA__GUGCGGCUGCCAGAAGUGGUUAC____UUCUGGGUGGCAACUUUGGCAAAUGGGCGCCUGGGA______CUGAGAGGGCAUCAGUUGGAUGG ...(((((((((((((((...(......)...)))))))(((((.(((.(((((..........))))).))))))))........((((................))))....)))))))). (-25.05 = -26.59 + 1.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:20 2011