
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,806,672 – 6,806,765 |
| Length | 93 |
| Max. P | 0.993082 |

| Location | 6,806,672 – 6,806,765 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.65389 |
| G+C content | 0.44775 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -13.60 |
| Energy contribution | -14.98 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
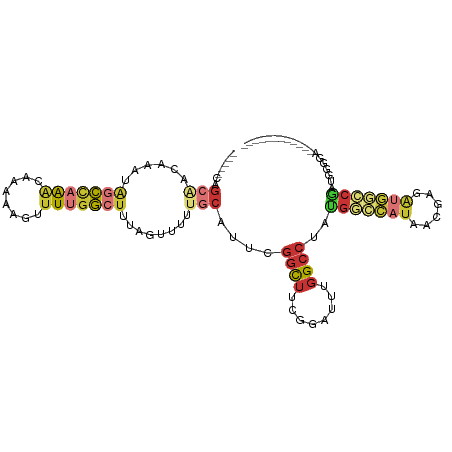
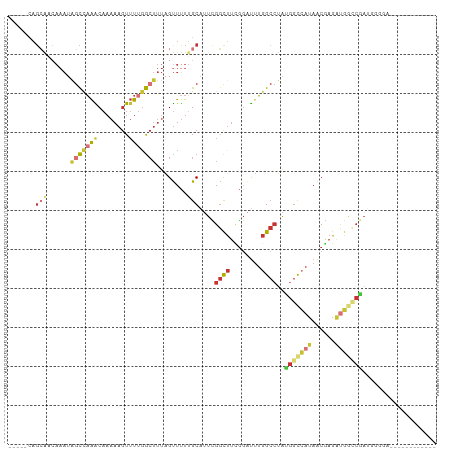
>dm3.chrX 6806672 93 + 22422827 -----CAGCAACAAAUAGCCAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGCUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUUUCGA------------ -----..((((.((..(((((((........)))))))....)).))))..(((((((((....(((((...)))))...)))...))))))......------------ ( -31.50, z-score = -3.36, R) >droSim1.chrX 5336990 89 + 17042790 -----CAGCAACAAAUAGCCAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGCUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUU---------------- -----..((((.((..(((((((........)))))))....)).))))..(((((((((....(((((...)))))...)))...))))))..---------------- ( -31.50, z-score = -3.79, R) >droSec1.super_24 20210 105 + 912625 -----CAGCAACAAAUAGCCAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGCUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUUGGGCUGGGAGGGUUGA -----..((((.((..(((((((........)))))))....)).))))..(((((((....((..((((((((((((......)))))))..)))))..)).))))))) ( -36.40, z-score = -2.55, R) >droYak2.chrX 7223123 101 - 21770863 -----CAGCAGCAAAUAGCUAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGCUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUAGGAACGGGUUGA---- -----((((.(((((.((((((.((((....))))...)))))))))))..(((((((((....(((((...)))))...)))...)))))).........)))).---- ( -32.70, z-score = -2.48, R) >droEre2.scaffold_4690 15732422 101 + 18748788 -----CAGCAACAAAUAGCCAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGUUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUGCGGAUGGGUUGA---- -----(((((((....(((((((........)))))))...)))(((((((.((((((((....(((((...)))))...))))...)))))))))))...)))).---- ( -33.90, z-score = -2.64, R) >droAna3.scaffold_13117 1651517 99 - 5790199 -----AAGCG--AAAUGGCUAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUUGGCUUUGGAUUUGGCCUAAGGCCAUAACGAGAUGAGACAAGGGGGGGGAAGAA---- -----..(((--(((.(((((((........))))))).....))))))...(((((((((.......))))))))).............................---- ( -22.20, z-score = -0.16, R) >droWil1.scaffold_180777 2890774 74 + 4753960 ----------------AGUCAGGCAAAAACUAUACAAAAAAGUUUUGGC-UUUGGCUUUGCCUUUGGCCUAUGGCAAUAACAAGGCUUACA------------------- ----------------((((((((.((((((.........)))))).))-.))))))..(((((..(((...)))......))))).....------------------- ( -18.10, z-score = -0.37, R) >droVir3.scaffold_12928 7543647 98 - 7717345 UGAGGCAGGGCCAAA-AACUCAGCAAAAAGUUUUGGCUUUGGCUCUGGCUGUUGCCCCAAGGCAGCGAACGGGCUAAAAGCGAGCCGAGCCAGGUGAAA----------- .(((.((((((((((-(.((........))))))))))))).)))(((((((((((....)))))).....((((.......)))).))))).......----------- ( -41.20, z-score = -1.85, R) >consensus _____CAGCAACAAAUAGCCAAACAAAAAGUUUUGGCUUUAGUUUUUGCAUUCGGCUUCGGAUUUGGCCUAUGGCCAUAACGAGAUGGCCGAUGGGGA____________ .......(((......(((((((........)))))))........)))....((((........))))..(((((((......)))))))................... (-13.60 = -14.98 + 1.38)

| Location | 6,806,672 – 6,806,765 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.65389 |
| G+C content | 0.44775 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.21 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
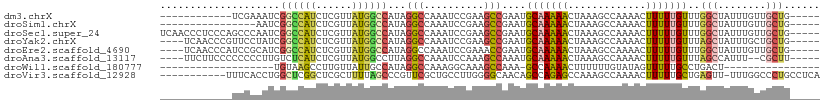
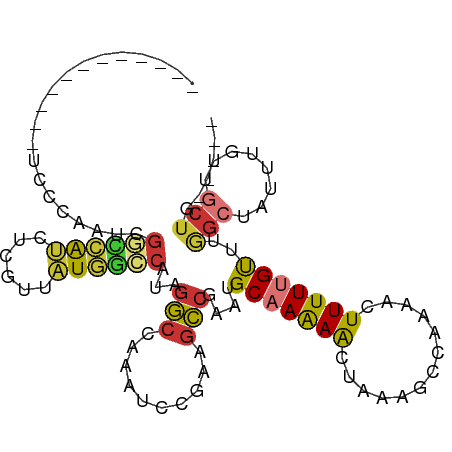
>dm3.chrX 6806672 93 - 22422827 ------------UCGAAAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAGCCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUGGCUAUUUGUUGCUG----- ------------......(((((....(((...(((((...)))))....))).)))))..((((.......(((((((.(.....)))))))).....))))..----- ( -27.00, z-score = -2.71, R) >droSim1.chrX 5336990 89 - 17042790 ----------------AAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAGCCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUGGCUAUUUGUUGCUG----- ----------------..(((((....(((...(((((...)))))....))).)))))..((((.......(((((((.(.....)))))))).....))))..----- ( -27.00, z-score = -2.99, R) >droSec1.super_24 20210 105 - 912625 UCAACCCUCCCAGCCCAAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAGCCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUGGCUAUUUGUUGCUG----- ..........((((.((.(((((....(((...(((((...)))))....))).))))).))....(((...(((((((.(.....))))))))....)))))))----- ( -28.90, z-score = -3.00, R) >droYak2.chrX 7223123 101 + 21770863 ----UCAACCCGUUCCUAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAGCCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUAGCUAUUUGCUGCUG----- ----..........(((((.((((((......)))))))))))..........((((((((((.(((...((((......))))...))).)).)))))..))).----- ( -22.70, z-score = -1.58, R) >droEre2.scaffold_4690 15732422 101 - 18748788 ----UCAACCCAUCCGCAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAACCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUGGCUAUUUGUUGCUG----- ----.((((......(((((((.....(((...(((((...)))))....)))..))).)))).........(((((((.(.....))))))))....))))...----- ( -26.20, z-score = -2.50, R) >droAna3.scaffold_13117 1651517 99 + 5790199 ----UUCUUCCCCCCCCUUGUCUCAUCUCGUUAUGGCCUUAGGCCAAAUCCAAAGCCAAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUAGCCAUUU--CGCUU----- ----.............................(((((...)))))......((((.(((((...((((.((((......))))..))))...)))))--.))))----- ( -16.00, z-score = -1.88, R) >droWil1.scaffold_180777 2890774 74 - 4753960 -------------------UGUAAGCCUUGUUAUUGCCAUAGGCCAAAGGCAAAGCCAAA-GCCAAAACUUUUUUGUAUAGUUUUUGCCUGACU---------------- -------------------.....(((((......(((...)))..))))).........-((.((((((.........)))))).))......---------------- ( -13.40, z-score = 0.70, R) >droVir3.scaffold_12928 7543647 98 + 7717345 -----------UUUCACCUGGCUCGGCUCGCUUUUAGCCCGUUCGCUGCCUUGGGGCAACAGCCAGAGCCAAAGCCAAAACUUUUUGCUGAGUU-UUUGGCCCUGCCUCA -----------.......((((((((((.((.....))......(.((((....)))).))))).))))))..((((((((((......))).)-))))))......... ( -32.60, z-score = -0.50, R) >consensus ____________UCCCAAUCGGCCAUCUCGUUAUGGCCAUAGGCCAAAUCCGAAGCCGAAUGCAAAAACUAAAGCCAAAACUUUUUGUUUGGCUAUUUGUUGCUG_____ ....................((((((......))))))...(((..........)))....(((((((.............)))))))..(((........)))...... ( -9.24 = -9.21 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:16 2011