
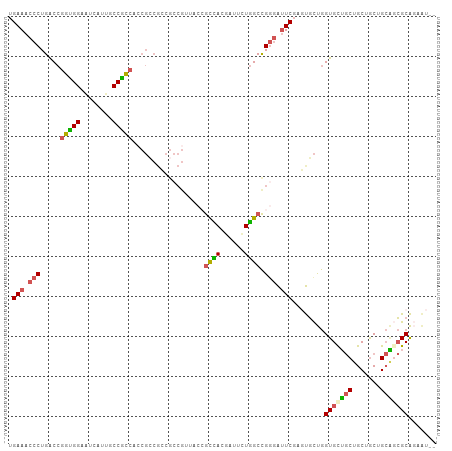
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,736,886 – 6,737,059 |
| Length | 173 |
| Max. P | 0.889818 |

| Location | 6,736,886 – 6,736,991 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.19 |
| Shannon entropy | 0.57282 |
| G+C content | 0.64250 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.72 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6736886 105 + 22422827 UGAAACCCUGACCGGUGGUAUCAUUGCCGCCACCGCCGCCGCCGUUACCGCCACGAUUCUGGCCGGGAUUCGAGUGCUGGUGUUGCUGCUGCUGCAGCGCAGAAU-- .......(((...(((((((....)))))))..(((((.(((((...((((((......))).)))....)).))).)))))..(((((....))))).)))...-- ( -41.30, z-score = -1.13, R) >droSim1.chrX_random 2160843 105 + 5698898 UGAAACCCUGACCGGCGGAAUCAUUGCCGCCAGCGCCGCCGCCGUUACCGCCACGAUUCUGGCCGGGAUUCGAGUGCUGGUGCUGCUGCUGCUGCAGCGCAGAAU-- .......(((...(((((........)))))(((((((.(((((...((((((......))).)))....)).))).)))))))(((((....))))).)))...-- ( -45.00, z-score = -1.55, R) >droSec1.super_4 6120154 105 - 6179234 UGAAACCCUGACCGGCGGAAUCAUUGCCGCCACCGCCGCCGCCGUUACCGCCACGAUUCUGGCCGGGAUUCGAGUGCUGGUGCUGCUGCUGCUGCAGCGCAGAAU-- .......(((...(((((........)))))..(((((.(((((...((((((......))).)))....)).))).)))))..(((((....))))).)))...-- ( -39.70, z-score = -0.49, R) >droYak2.chrX 2703782 81 + 21770863 UGAAACCCUGACCGCUGGAGUCAUUGCCGGC------------------GUUACGAUUUUGGCCGGGAUUC---UGUUGGUGCUGCUGCUGCAGCGCCGCAU----- .(((.(((.....(((((........)))))------------------((((......)))).))).)))---(((.((((((((....))))))))))).----- ( -31.70, z-score = -1.41, R) >droWil1.scaffold_180777 2851743 102 - 4753960 UGAAACCCUGGCCAGUGGCAUCGCCACCACCGCCGACGGCGCCG-----ACGAUGCCUGUCGCUGAUGCUCCUCCUCCAGUGCGACCGCCACCGGCCGGCGAUGUGU .....(.((((((.(((((.((((..........(((((.((..-----.....)))))))((((..(......)..))))))))..))))).)))))).)...... ( -37.20, z-score = -0.22, R) >consensus UGAAACCCUGACCGGUGGAAUCAUUGCCGCCACCGCCGCCGCCGUUACCGCCACGAUUCUGGCCGGGAUUCGAGUGCUGGUGCUGCUGCUGCUGCAGCGCAGAAU__ .(((.(((.....(((((........)))))..................((((......)))).))).)))........(((((((.......)))))))....... (-23.76 = -23.72 + -0.04)


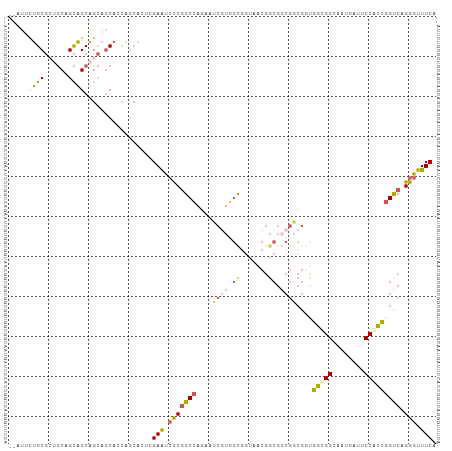
| Location | 6,736,886 – 6,736,991 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.19 |
| Shannon entropy | 0.57282 |
| G+C content | 0.64250 |
| Mean single sequence MFE | -41.41 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6736886 105 - 22422827 --AUUCUGCGCUGCAGCAGCAGCAACACCAGCACUCGAAUCCCGGCCAGAAUCGUGGCGGUAACGGCGGCGGCGGUGGCGGCAAUGAUACCACCGGUCAGGGUUUCA --.......(((((....))))).............(((.(((((((...(((((.((.((....)).)).)))))((.((........)).)))))).))).))). ( -38.90, z-score = 0.15, R) >droSim1.chrX_random 2160843 105 - 5698898 --AUUCUGCGCUGCAGCAGCAGCAGCACCAGCACUCGAAUCCCGGCCAGAAUCGUGGCGGUAACGGCGGCGGCGCUGGCGGCAAUGAUUCCGCCGGUCAGGGUUUCA --....((((((((....))))).))).........(((.(((((((.....(((.((.((....)).)).)))..(((((........))))))))).))).))). ( -43.00, z-score = -0.05, R) >droSec1.super_4 6120154 105 + 6179234 --AUUCUGCGCUGCAGCAGCAGCAGCACCAGCACUCGAAUCCCGGCCAGAAUCGUGGCGGUAACGGCGGCGGCGGUGGCGGCAAUGAUUCCGCCGGUCAGGGUUUCA --....((((((((....))))).))).........(((.(((((((...(((((.((.((....)).)).)))))(((((........))))))))).))).))). ( -45.90, z-score = -0.98, R) >droYak2.chrX 2703782 81 - 21770863 -----AUGCGGCGCUGCAGCAGCAGCACCAACA---GAAUCCCGGCCAAAAUCGUAAC------------------GCCGGCAAUGACUCCAGCGGUCAGGGUUUCA -----....((.(((((....))))).))....---(((.(((.(((...........------------------...)))..(((((.....)))))))).))). ( -27.64, z-score = -1.03, R) >droWil1.scaffold_180777 2851743 102 + 4753960 ACACAUCGCCGGCCGGUGGCGGUCGCACUGGAGGAGGAGCAUCAGCGACAGGCAUCGU-----CGGCGCCGUCGGCGGUGGUGGCGAUGCCACUGGCCAGGGUUUCA ........(((((((((....(((((..((..(......)..))))))).((((((((-----((.(((((....))))).))))))))))))))))).))...... ( -51.60, z-score = -2.00, R) >consensus __AUUCUGCGCUGCAGCAGCAGCAGCACCAGCACUCGAAUCCCGGCCAGAAUCGUGGCGGUAACGGCGGCGGCGGUGGCGGCAAUGAUUCCACCGGUCAGGGUUUCA .....((((......)))).................(((.(((((((....((((.((.......)).))))....(((((........))))))))).))).))). (-22.54 = -23.26 + 0.72)


| Location | 6,736,954 – 6,737,059 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 63.05 |
| Shannon entropy | 0.75578 |
| G+C content | 0.65051 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.81 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6736954 105 + 22422827 UUCGAGUGCUGGUGUUGCUGCUGCUG--CAGCGCAGAAUC---GGCGGCUCCUGUUGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUCGCCGGA----- (((..((((((.....((....))..--)))))).)))((---(((((((...........((((.((......)).))))((((.(......).)))).))))))))).----- ( -42.20, z-score = -0.30, R) >droSim1.chrX_random 2160911 105 + 5698898 UUCGAGUGCUGGUGCUGCUGCUGCUG--CAGCGCAGAAUC---GGCGGCUCCUGUCGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUCGCCGGG----- ...((((((((((.((((.((((...--)))))))).)))---))).))))(((.(((.((((((.((......)).))))((((.(......).)))).))))).))).----- ( -44.90, z-score = -0.49, R) >droSec1.super_4 6120222 105 - 6179234 UUCGAGUGCUGGUGCUGCUGCUGCUG--CAGCGCAGAAUC---GGCGGCUCCUGUCGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUCGCCGGG----- ...((((((((((.((((.((((...--)))))))).)))---))).))))(((.(((.((((((.((......)).))))((((.(......).)))).))))).))).----- ( -44.90, z-score = -0.49, R) >droYak2.chrX 2703832 99 + 21770863 ------UUCUGUUGGUGCUGCUGCUG--CAGCGCCGCAUC---AGCGGCUCCUGUCGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUAGCCGGA----- ------..(((.(((((((((....)--))))))))...)---))(((((((.........((((.((......)).))))((((.(......).)))).)).)))))..----- ( -45.80, z-score = -1.83, R) >droEre2.scaffold_4690 15664504 102 + 18748788 UUUGAAUGCUGGUGCUGCUGCUG-----CAGCGCAGAAUC---GGCGGCUCCUGUCGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUCGCCGGA----- ...........(((((((....)-----))))))....((---(((((((...........((((.((......)).))))((((.(......).)))).))))))))).----- ( -44.50, z-score = -1.65, R) >droAna3.scaffold_13117 536319 101 + 5790199 ------UGCGACUGGUGGUCACAUUGGGAGCAGUCGGCUC---CGCCGCUCCCGUUGAGCUGCUGGAAAAGGUCCUGGAGCGUGGGCUCUGCUGGCCGCUAGUGGCUGGC----- ------.((.((((((((((.((.(((((((.(.((....---))).))))))).))(((....(((.....))).(((((....)))))))))))))))))).))....----- ( -48.80, z-score = -1.44, R) >droWil1.scaffold_180777 2851806 110 - 4753960 CUCCUCCUCCAGUGCGACCGCCACCGGCCGGCGAUGUGUCCACAGCGGCUCCAGUUGAGCUGCUGCUAAAUGUACGAGAGCGUGGGCUCUGUUGGCCACUGGUCGCUGGA----- .......((((..(((((((.....((((((((..(.((((((((((((((.....))))))))(((...........))))))))).)))))))))..)))))))))))----- ( -50.40, z-score = -1.98, R) >dp4.chrXL_group1a 8722516 84 + 9151740 -----------------------CCCGGUGGCGAUGUGUC---AGCGGCUCCCGUGGAGCUGCUCGAGAAUGUCCUGGAGCGCGGGCUCUGCUGGCCACUGGUCGCCGGU----- -----------------------.((((((((.(((.(((---(((((..(((((......((((.((......)).)))))))))..)))))))))).).)))))))).----- ( -45.40, z-score = -2.46, R) >droPer1.super_15 1118956 84 + 2181545 -----------------------CCCGGUGGCGAUGUGUC---AGGGGCUCCCGUGGAGCUGCUCGAGAAUGUCCUGGAGCGCGGGCUCUGCUGGCCACUGGUCGCCGGU----- -----------------------.((((((((.(((.(((---((((...)))(..(((((((((.((......)).))))...)))))..))))))).).)))))))).----- ( -41.70, z-score = -1.26, R) >anoGam1.chrX 6810202 90 + 22145176 ------------------------UCAGUCAGGUAGGUUU-GCUGCGCUGGCAGCGAUUCGGCCCAAAGCCGAUGGUGGUGGUAGCCACCCAUGCCAUGCGGCAGGUGGGCCGAA ------------------------...(((((((((....-.)))).))))).....(((((((((..((((((((((((((...)))))...))))).))))...))))))))) ( -48.20, z-score = -3.46, R) >consensus ______UGCUGGUGCUGCUGCUGCUG__CAGCGCAGAAUC___AGCGGCUCCUGUCGAACUGCUCGAGAAUGUCCUGGAGCGUGGGCUCUGUUGGCCACUGGUCGCCGGA_____ .............................................((((..(((....((.((((.(........).)))))).((((.....))))..)))..))))....... (-13.59 = -13.81 + 0.22)


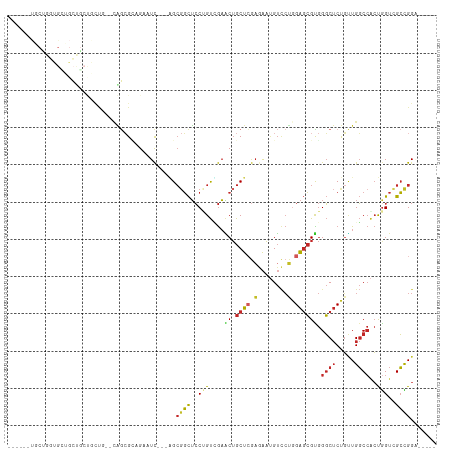
| Location | 6,736,954 – 6,737,059 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 63.05 |
| Shannon entropy | 0.75578 |
| G+C content | 0.65051 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -13.08 |
| Energy contribution | -14.17 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6736954 105 - 22422827 -----UCCGGCGACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCAACAGGAGCCGCC---GAUUCUGCGCUG--CAGCAGCAGCAACACCAGCACUCGAA -----..(((((....((((.(......).))))((((.(((....))).)))).((((.....))))))))---).......((((--(....)))))................ ( -32.80, z-score = -0.64, R) >droSim1.chrX_random 2160911 105 - 5698898 -----CCCGGCGACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCGACAGGAGCCGCC---GAUUCUGCGCUG--CAGCAGCAGCAGCACCAGCACUCGAA -----..(((((....((((.(......).))))(((((.((((...........))))....)))))))))---)....(((((((--(....))))).)))............ ( -36.00, z-score = -0.79, R) >droSec1.super_4 6120222 105 + 6179234 -----CCCGGCGACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCGACAGGAGCCGCC---GAUUCUGCGCUG--CAGCAGCAGCAGCACCAGCACUCGAA -----..(((((....((((.(......).))))(((((.((((...........))))....)))))))))---)....(((((((--(....))))).)))............ ( -36.00, z-score = -0.79, R) >droYak2.chrX 2703832 99 - 21770863 -----UCCGGCUACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCGACAGGAGCCGCU---GAUGCGGCGCUG--CAGCAGCAGCACCAACAGAA------ -----((((((((....)))))....((((....((((.(((....))).)))).))))....)))(((((.---...)))))((((--(....)))))..........------ ( -39.90, z-score = -2.36, R) >droEre2.scaffold_4690 15664504 102 - 18748788 -----UCCGGCGACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCGACAGGAGCCGCC---GAUUCUGCGCUG-----CAGCAGCAGCACCAGCAUUCAAA -----..(((((....((((.(......).))))(((((.((((...........))))....)))))))))---)...(((.((((-----(....)))))..)))........ ( -34.30, z-score = -1.24, R) >droAna3.scaffold_13117 536319 101 - 5790199 -----GCCAGCCACUAGCGGCCAGCAGAGCCCACGCUCCAGGACCUUUUCCAGCAGCUCAACGGGAGCGGCG---GAGCCGACUGCUCCCAAUGUGACCACCAGUCGCA------ -----((..(((......)))..)).((((....(((...(((.....)))))).))))...((((((((((---....)).))))))))..((((((.....))))))------ ( -40.50, z-score = -2.14, R) >droWil1.scaffold_180777 2851806 110 + 4753960 -----UCCAGCGACCAGUGGCCAACAGAGCCCACGCUCUCGUACAUUUAGCAGCAGCUCAACUGGAGCCGCUGUGGACACAUCGCCGGCCGGUGGCGGUCGCACUGGAGGAGGAG -----((((((((((.(((.((.(((((((....))))).)).......(((((.((((.....)))).))))))).))).(((((....))))).))))))..))))....... ( -47.70, z-score = -1.67, R) >dp4.chrXL_group1a 8722516 84 - 9151740 -----ACCGGCGACCAGUGGCCAGCAGAGCCCGCGCUCCAGGACAUUCUCGAGCAGCUCCACGGGAGCCGCU---GACACAUCGCCACCGGG----------------------- -----...(((((...(((..((((.........((((.(((....))).)))).(((((...))))).)))---).))).)))))......----------------------- ( -32.40, z-score = -0.94, R) >droPer1.super_15 1118956 84 - 2181545 -----ACCGGCGACCAGUGGCCAGCAGAGCCCGCGCUCCAGGACAUUCUCGAGCAGCUCCACGGGAGCCCCU---GACACAUCGCCACCGGG----------------------- -----...(((((...(((..(((..(..(((((((((.(((....))).)))).)).....)))..)..))---).))).)))))......----------------------- ( -28.00, z-score = 0.11, R) >anoGam1.chrX 6810202 90 - 22145176 UUCGGCCCACCUGCCGCAUGGCAUGGGUGGCUACCACCACCAUCGGCUUUGGGCCGAAUCGCUGCCAGCGCAGC-AAACCUACCUGACUGA------------------------ (((((((((...((((.((((....(((((...))))).))))))))..)))))))))..(((((....)))))-................------------------------ ( -44.40, z-score = -4.46, R) >consensus _____UCCGGCGACCAGUGGCCAACAGAGCCCACGCUCCAGGACAUUCUCGAGCAGUUCGACAGGAGCCGCC___GAUUCUGCGCUG__CAGCAGCAGCAGCACCAGCA______ ........((((....((((.(......).))))((((.((......)).)))).((((.....))))))))........................................... (-13.08 = -14.17 + 1.09)

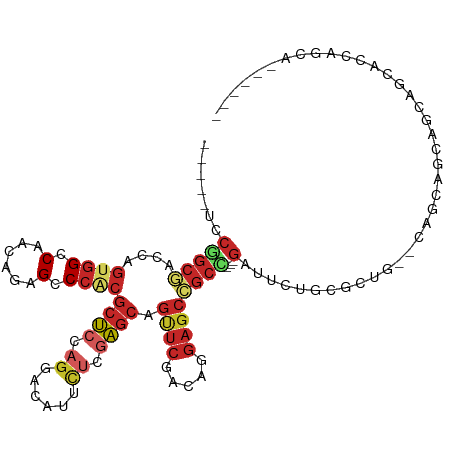

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:05 2011