
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,732,906 – 6,732,999 |
| Length | 93 |
| Max. P | 0.939229 |

| Location | 6,732,906 – 6,732,999 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.96 |
| Shannon entropy | 0.16383 |
| G+C content | 0.42373 |
| Mean single sequence MFE | -16.98 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
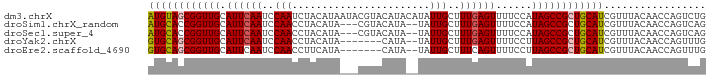
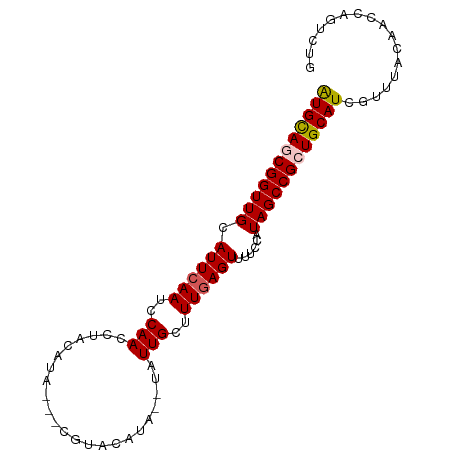
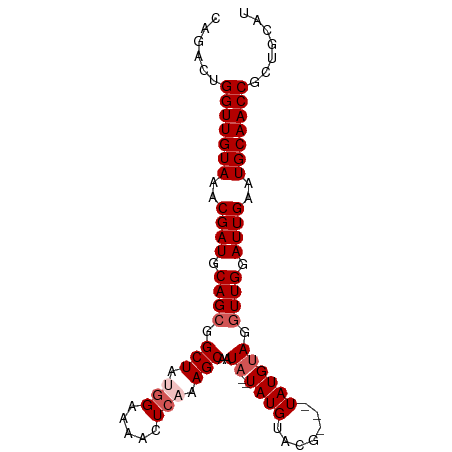
>dm3.chrX 6732906 93 + 22422827 AUGUAGCGGUUGCAUUCAAUCCAAUCUACAUAAUACGUACAUACAUAUUGCUUUGAGUUUUCCAUAGCCGCUGCAUCGUUUACAACCAGUCUG ((((((((((((.((((((..((((.....................))))..))))))......))))))))))))................. ( -18.90, z-score = -1.65, R) >droSim1.chrX_random 2156569 88 + 5698898 AUGCACCGGUUGCAUUCAAUCCAACCUACAUA---CGUACAUA--UAUUGCUUUGAGUUUUCCAUAGCCGCUGCAUCGUUUACAACCAGUCAG (((((.((((((.((((((..(((..(((...---.)))....--..)))..))))))......)))))).)))))................. ( -14.20, z-score = -0.68, R) >droSec1.super_4 6115990 88 - 6179234 AUGCACCGGUUGCAUUCAAUCCAACCUACAUA---CGUACAUA--UAUUGCUUUGAGUUUUCCAUAGCCGCUGCAUCGUUUACAACCAGUCAG (((((.((((((.((((((..(((..(((...---.)))....--..)))..))))))......)))))).)))))................. ( -14.20, z-score = -0.68, R) >droYak2.chrX 2700142 84 + 21770863 GUGCAGCGGUUGCAUUCAAUCCAACCUACAUA-------CAUA--UAUUGCUUUGAGUUUUCCUUAGCCGCUGCAUCGUUUACAACCAGUUUG ((((((((((((.((((((..(((..((....-------..))--..)))..))))))......))))))))))))................. ( -21.20, z-score = -3.01, R) >droEre2.scaffold_4690 15660633 84 + 18748788 GUGCAGCGGUUGCAUUCAAUCCAACCUUCAUA-------CAUA--UAUUGCUUUCAGUUUUCCUUAGCCGCUGCAUCGUUUACAACCAGUUUG ((((((((((((((..................-------....--...)))....((.....)).)))))))))))................. ( -16.41, z-score = -2.10, R) >consensus AUGCAGCGGUUGCAUUCAAUCCAACCUACAUA___CGUACAUA__UAUUGCUUUGAGUUUUCCAUAGCCGCUGCAUCGUUUACAACCAGUCUG ((((((((((((.((((((..(((.......................)))..))))))......))))))))))))................. (-16.26 = -16.46 + 0.20)
| Location | 6,732,906 – 6,732,999 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.96 |
| Shannon entropy | 0.16383 |
| G+C content | 0.42373 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
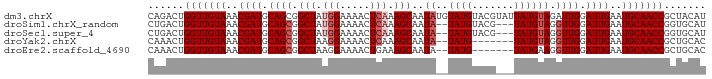
>dm3.chrX 6732906 93 - 22422827 CAGACUGGUUGUAAACGAUGCAGCGGCUAUGGAAAACUCAAAGCAAUAUGUAUGUACGUAUUAUGUAGAUUGGAUUGAAUGCAACCGCUACAU ......(((((((..((((.((((.(((.(((.....))).)))(((((((....))))))).....).))).))))..)))))))....... ( -22.80, z-score = -1.29, R) >droSim1.chrX_random 2156569 88 - 5698898 CUGACUGGUUGUAAACGAUGCAGCGGCUAUGGAAAACUCAAAGCAAUA--UAUGUACG---UAUGUAGGUUGGAUUGAAUGCAACCGGUGCAU .((((((((((((..((((.((((.(((.(((.....))).))).(((--(((....)---)))))..)))).))))..)))))))))).)). ( -29.90, z-score = -3.73, R) >droSec1.super_4 6115990 88 + 6179234 CUGACUGGUUGUAAACGAUGCAGCGGCUAUGGAAAACUCAAAGCAAUA--UAUGUACG---UAUGUAGGUUGGAUUGAAUGCAACCGGUGCAU .((((((((((((..((((.((((.(((.(((.....))).))).(((--(((....)---)))))..)))).))))..)))))))))).)). ( -29.90, z-score = -3.73, R) >droYak2.chrX 2700142 84 - 21770863 CAAACUGGUUGUAAACGAUGCAGCGGCUAAGGAAAACUCAAAGCAAUA--UAUG-------UAUGUAGGUUGGAUUGAAUGCAACCGCUGCAC ......(((((((..((((.((((.(((...((....))..)))..((--((..-------..)))).)))).))))..)))))))....... ( -20.30, z-score = -1.08, R) >droEre2.scaffold_4690 15660633 84 - 18748788 CAAACUGGUUGUAAACGAUGCAGCGGCUAAGGAAAACUGAAAGCAAUA--UAUG-------UAUGAAGGUUGGAUUGAAUGCAACCGCUGCAC .......((((....))))(((((.(((.((.....))...)))....--....-------......(((((.((...)).)))))))))).. ( -20.50, z-score = -1.83, R) >consensus CAGACUGGUUGUAAACGAUGCAGCGGCUAUGGAAAACUCAAAGCAAUA__UAUGUACG___UAUGUAGGUUGGAUUGAAUGCAACCGCUGCAU ......(((((((..((((.((((.(((.(((.....))).)))......((((.......))))...)))).))))..)))))))....... (-17.42 = -18.22 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:22:01 2011