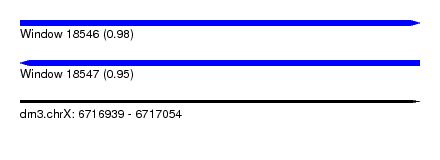
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,716,939 – 6,717,054 |
| Length | 115 |
| Max. P | 0.983982 |

| Location | 6,716,939 – 6,717,054 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 61.78 |
| Shannon entropy | 0.78702 |
| G+C content | 0.57710 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.64 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
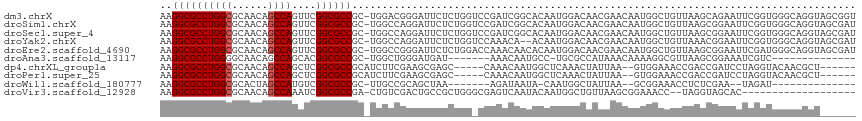
>dm3.chrX 6716939 115 + 22422827 AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC-UGGACGGGAUUCUCUGGUCCGAUCGGCACAAUGGACAACGAACAAUGGCUGUUAAGCAGAAUUCGGUGGGCAGGUAGCGGU ...(((((((.((((((((((((((((..((((.-(((((.((.....)).)))))..)))).(....)....)))))..))))))))..)).(....)....).))))).))... ( -44.70, z-score = -1.17, R) >droSim1.chrX 5271859 115 + 17042790 AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC-UGGCCAGGAUUCUCUGGUCCGAUCGGCACAAUGGACAACGAACAAUGGCUGUUAAGCGGAAUUCGGUGGGCAGGUAGCGAU ...(((((((.(..(((((((((((((..((((.-(((((.((((......))))....))).)).))).)..)))))..))))))))..((.(....).)).).))))).))... ( -44.90, z-score = -0.94, R) >droSec1.super_4 6100052 115 - 6179234 AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC-UGGCCAGGAUUCUCUGGUCCGAUCGGCACAAUGGACAACGAACAAUGGCUGUUAAGCGGAAUUCGGUGGGCAGGUAGCGAU ...(((((((.(..(((((((((((((..((((.-(((((.((((......))))....))).)).))).)..)))))..))))))))..((.(....).)).).))))).))... ( -44.90, z-score = -0.94, R) >droYak2.chrX 2684420 113 + 21770863 AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC-UGGCCAGGAUUCUCUGGUCCAAACA--ACAAUGGACAACGAACAAUGGCUGUUAAACGGAAUUCGGUGGGCAGGUAGCGAU ..((((((((((......))))....))))))((-(.(((.(((((((...(((((....--....)))))....((((.....))))....)))))))))).))).......... ( -43.20, z-score = -1.87, R) >droEre2.scaffold_4690 15644880 115 + 18748788 AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC-UGGCCGGGAUUCUCUGGACCAAACAACACAAUGGACAACGAACAAUGGCUGUUAAGCGGAAUUCGAUGGGCAGGUAGCGAU ..((((((((((......))))....))))))((-((((((((....))))).)).....((....((..((.((((...(.(((....))).)..)))).))..)).)))))... ( -39.50, z-score = -0.72, R) >droAna3.scaffold_13117 516874 93 + 5790199 AAGGCGCCUGGGGCAACAGCCAGCACGGCGCCGC-UGGCUGGGAUGAU-------AAACAAUGCC-UGCGCCAUAAACAAAAGGCGUUAAGCGGAAAUCGUC-------------- ..(((((...(((((.((((((((........))-))))))...((..-------...)).))))-))))))...........((.....))..........-------------- ( -33.70, z-score = -0.70, R) >dp4.chrXL_group1a 6546084 103 + 9151740 AAGGCGCCUGGCGCAACAGCCAGCUCGGCGCCGCAUCUUCGAAGCGAGC-----CAAACAAUGGCUCAAACUAUUAA--GUGGAAACCGACCGAUCCUAGGUACAACGCU------ ..((((((((((......))))....))))))((.((...)).))((((-----((.....)))))).........(--(((.......(((.......)))....))))------ ( -35.20, z-score = -1.81, R) >droPer1.super_25 680749 103 - 1448063 AAGGCGCCUGGCGCAACAGCCAGCUCGGCGCCGCAUCUUCGAAGCGAGC-----CAAACAAUGGCUCAAACUAUUAA--GUGGAAACCGACCGAUCCUAGGUACAACGCU------ ..((((((((((......))))....))))))((.((...)).))((((-----((.....)))))).........(--(((.......(((.......)))....))))------ ( -35.20, z-score = -1.81, R) >droWil1.scaffold_180777 1900099 89 + 4753960 AAGGCGCCUGGCGCACUAGCCAUGUCGGCGCCGC-UUGCCGCAGCUAA-------AGAUAAUA-CAAUGGCUAUUAA--GCGGAAACCUCUCGAA--UAGAU-------------- ..((((((((((......))))....))))))..-...(((((((((.-------.(......-)..))))).....--))))............--.....-------------- ( -27.40, z-score = -0.26, R) >droVir3.scaffold_12928 3697743 94 - 7717345 AAGGCGCCUGGCGCAACAGCCAAAUCGGCGCCGA-CUGUCGACUGCCGCUGGGCGAGUCAAUACAAUGGCUGUUAAGCGGAAACC--UAGGUAGCAC------------------- ..((((((((((......))))....))))))..-.....(.((((((((..((.(((((......)))))))..)))(....).--..))))))..------------------- ( -38.60, z-score = -1.97, R) >consensus AAGGCGCCUGGCGCAACAGCCAGUUCGGCGCCGC_UGGCCGGGAUUCUCUGG_CCAAACAAUACAAUGGACAACGAACAAUGGCUGUUAAGCGGAAUUCGGUGGGCAGGU______ ..((((((((((......))))....)))))).................................................................................... (-18.54 = -18.64 + 0.10)

| Location | 6,716,939 – 6,717,054 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 61.78 |
| Shannon entropy | 0.78702 |
| G+C content | 0.57710 |
| Mean single sequence MFE | -36.39 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
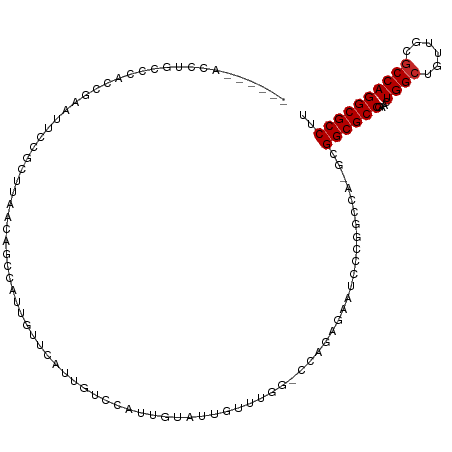
>dm3.chrX 6716939 115 - 22422827 ACCGCUACCUGCCCACCGAAUUCUGCUUAACAGCCAUUGUUCGUUGUCCAUUGUGCCGAUCGGACCAGAGAAUCCCGUCCA-GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ...((..((((......(....).((.(((((((((..(((((.........((((((...((((..(.....)..)))).-.)))))))))))))))))))).)))))).))... ( -38.40, z-score = -1.60, R) >droSim1.chrX 5271859 115 - 17042790 AUCGCUACCUGCCCACCGAAUUCCGCUUAACAGCCAUUGUUCGUUGUCCAUUGUGCCGAUCGGACCAGAGAAUCCUGGCCA-GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ...((..((((......(.....)((.(((((((((..(((((.........((((((...((.((((......)))))).-.)))))))))))))))))))).)))))).))... ( -42.10, z-score = -1.94, R) >droSec1.super_4 6100052 115 + 6179234 AUCGCUACCUGCCCACCGAAUUCCGCUUAACAGCCAUUGUUCGUUGUCCAUUGUGCCGAUCGGACCAGAGAAUCCUGGCCA-GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ...((..((((......(.....)((.(((((((((..(((((.........((((((...((.((((......)))))).-.)))))))))))))))))))).)))))).))... ( -42.10, z-score = -1.94, R) >droYak2.chrX 2684420 113 - 21770863 AUCGCUACCUGCCCACCGAAUUCCGUUUAACAGCCAUUGUUCGUUGUCCAUUGU--UGUUUGGACCAGAGAAUCCUGGCCA-GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ...(((.........((((((..((....(((((........)))))....)).--.)))))).((((......))))..)-))((((((....((((......)))))))))).. ( -36.90, z-score = -1.19, R) >droEre2.scaffold_4690 15644880 115 - 18748788 AUCGCUACCUGCCCAUCGAAUUCCGCUUAACAGCCAUUGUUCGUUGUCCAUUGUGUUGUUUGGUCCAGAGAAUCCCGGCCA-GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ...((..((((......(.....)((.(((((((((..(((((.........((((((((.((((..(.....)..)))))-))))))))))))))))))))).)))))).))... ( -35.90, z-score = -0.41, R) >droAna3.scaffold_13117 516874 93 - 5790199 --------------GACGAUUUCCGCUUAACGCCUUUUGUUUAUGGCGCA-GGCAUUGUUU-------AUCAUCCCAGCCA-GCGGCGCCGUGCUGGCUGUUGCCCCAGGCGCCUU --------------(((((.....((.....))...)))))...(((((.-((((.((...-------..))...((((((-(((......))))))))).))))....))))).. ( -31.70, z-score = -0.61, R) >dp4.chrXL_group1a 6546084 103 - 9151740 ------AGCGUUGUACCUAGGAUCGGUCGGUUUCCAC--UUAAUAGUUUGAGCCAUUGUUUG-----GCUCGCUUCGAAGAUGCGGCGCCGAGCUGGCUGUUGCGCCAGGCGCCUU ------.((((........(((..........))).(--((...(((..((((((.....))-----)))))))...)))))))((((((....((((......)))))))))).. ( -36.80, z-score = -0.36, R) >droPer1.super_25 680749 103 + 1448063 ------AGCGUUGUACCUAGGAUCGGUCGGUUUCCAC--UUAAUAGUUUGAGCCAUUGUUUG-----GCUCGCUUCGAAGAUGCGGCGCCGAGCUGGCUGUUGCGCCAGGCGCCUU ------.((((........(((..........))).(--((...(((..((((((.....))-----)))))))...)))))))((((((....((((......)))))))))).. ( -36.80, z-score = -0.36, R) >droWil1.scaffold_180777 1900099 89 - 4753960 --------------AUCUA--UUCGAGAGGUUUCCGC--UUAAUAGCCAUUG-UAUUAUCU-------UUAGCUGCGGCAA-GCGGCGCCGACAUGGCUAGUGCGCCAGGCGCCUU --------------.(((.--....))).((((((((--.....(((.(..(-......).-------.).))))))).))-))((((((....((((......)))))))))).. ( -27.40, z-score = 0.31, R) >droVir3.scaffold_12928 3697743 94 + 7717345 -------------------GUGCUACCUA--GGUUUCCGCUUAACAGCCAUUGUAUUGACUCGCCCAGCGGCAGUCGACAG-UCGGCGCCGAUUUGGCUGUUGCGCCAGGCGCCUU -------------------(((((.....--((...))((.(((((((((.....((((((.(((....))))))))).((-(((....)))))))))))))).))..)))))... ( -35.80, z-score = -1.34, R) >consensus ______ACCUGCCCACCGAAUUCCGCUUAACAGCCAUUGUUCAUUGUCCAUUGUAUUGUUUGG_CCAGAGAAUCCCGGCCA_GCGGCGCCGAACUGGCUGUUGCGCCAGGCGCCUU ....................................................................................((((((....((((......)))))))))).. (-17.42 = -17.52 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:59 2011