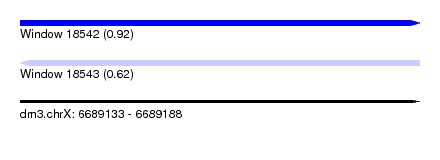
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,689,133 – 6,689,188 |
| Length | 55 |
| Max. P | 0.919611 |

| Location | 6,689,133 – 6,689,188 |
|---|---|
| Length | 55 |
| Sequences | 7 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.38812 |
| G+C content | 0.54071 |
| Mean single sequence MFE | -15.39 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.01 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
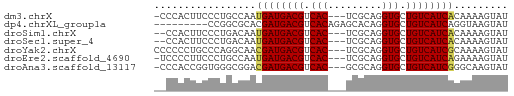
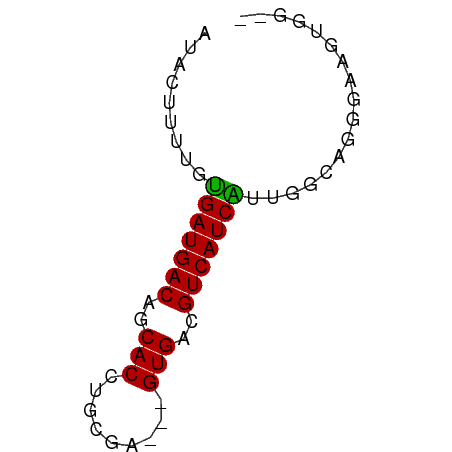
>dm3.chrX 6689133 55 + 22422827 -CCCACUUCCCUGCCAAUGAUGACGUCAC---UCGCAGGUGCUGUCAUCACAAAAGUAU -................((((((((.(((---(....)))).))))))))......... ( -13.90, z-score = -2.15, R) >dp4.chrXL_group1a 6514702 50 + 9151740 ---------CCGGCGCACGAUGACGUCACAGAGCACAGGUGCUGUCAUCAGGUAAGUAU ---------.....((..(((((((.(((.........))).)))))))..))...... ( -12.20, z-score = -0.06, R) >droSim1.chrX 5260715 54 + 17042790 --CCACUUCCCUGACAAUGAUGACGUCAC---UCGCAGGUGCUGUCAUCACAAAAGUAU --...............((((((((.(((---(....)))).))))))))......... ( -13.90, z-score = -1.88, R) >droSec1.super_4 6072925 54 - 6179234 --CCACUUCCCUGACAAUGAUGACGUCAC---UCGCAGGUGCUGUCAUCACAAAAGUAU --...............((((((((.(((---(....)))).))))))))......... ( -13.90, z-score = -1.88, R) >droYak2.chrX 2654924 56 + 21770863 CCCCCCUGCCCAGGCAACGAUGACGUCAC---UCGCAGGUGCUGUCAUCGCAAAAGUAU ......(((....))).((((((((.(((---(....)))).))))))))......... ( -18.00, z-score = -2.46, R) >droEre2.scaffold_4690 15616872 55 + 18748788 -UCCCCUUCCCUGCCAAUGAUGACGUCAC---UCGCAGGUGCUGUCAUCAGAAAAGUAU -.........((.....((((((((.(((---(....)))).))))))))....))... ( -14.50, z-score = -2.46, R) >droAna3.scaffold_13117 4481378 55 + 5790199 -CCCACCGGUGGGCGGACGAUGACGUCAC---GCGCAGGUGCUGUCAUCGGGCAAGUAU -((((....)))).(..((((((((.(((---......))).))))))))..)...... ( -21.30, z-score = -0.73, R) >consensus __CCACUUCCCUGCCAAUGAUGACGUCAC___UCGCAGGUGCUGUCAUCACAAAAGUAU .................((((((((.(((.........))).))))))))......... (-11.20 = -11.01 + -0.18)
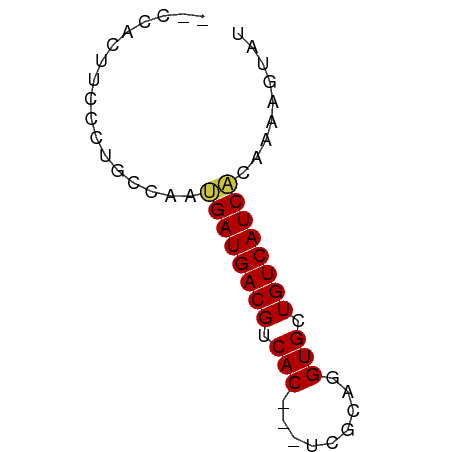
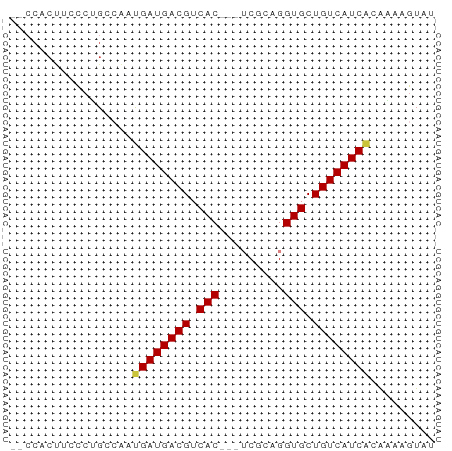
| Location | 6,689,133 – 6,689,188 |
|---|---|
| Length | 55 |
| Sequences | 7 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.38812 |
| G+C content | 0.54071 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
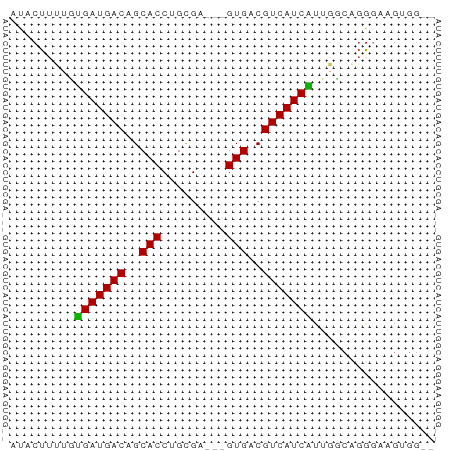
>dm3.chrX 6689133 55 - 22422827 AUACUUUUGUGAUGACAGCACCUGCGA---GUGACGUCAUCAUUGGCAGGGAAGUGGG- .(((((((((....)))...(((((.(---((((.....))))).)))))))))))..- ( -18.10, z-score = -2.02, R) >dp4.chrXL_group1a 6514702 50 - 9151740 AUACUUACCUGAUGACAGCACCUGUGCUCUGUGACGUCAUCGUGCGCCGG--------- ..........((((((((((....)))).......)))))).........--------- ( -10.31, z-score = 0.34, R) >droSim1.chrX 5260715 54 - 17042790 AUACUUUUGUGAUGACAGCACCUGCGA---GUGACGUCAUCAUUGUCAGGGAAGUGG-- .(((((((((((((((..(((......---)))..))))))))......))))))).-- ( -16.30, z-score = -1.42, R) >droSec1.super_4 6072925 54 + 6179234 AUACUUUUGUGAUGACAGCACCUGCGA---GUGACGUCAUCAUUGUCAGGGAAGUGG-- .(((((((((((((((..(((......---)))..))))))))......))))))).-- ( -16.30, z-score = -1.42, R) >droYak2.chrX 2654924 56 - 21770863 AUACUUUUGCGAUGACAGCACCUGCGA---GUGACGUCAUCGUUGCCUGGGCAGGGGGG ...(((((((((((((..(((......---)))..))))))))(((....)))))))). ( -18.90, z-score = -1.10, R) >droEre2.scaffold_4690 15616872 55 - 18748788 AUACUUUUCUGAUGACAGCACCUGCGA---GUGACGUCAUCAUUGGCAGGGAAGGGGA- ...((((((...((....))(((((.(---((((.....))))).)))))))))))..- ( -17.70, z-score = -2.00, R) >droAna3.scaffold_13117 4481378 55 - 5790199 AUACUUGCCCGAUGACAGCACCUGCGC---GUGACGUCAUCGUCCGCCCACCGGUGGG- .........(((((((..(((......---)))..))))))).(((((....))))).- ( -20.00, z-score = -1.36, R) >consensus AUACUUUUGUGAUGACAGCACCUGCGA___GUGACGUCAUCAUUGGCAGGGAAGUGG__ .........(((((((..(((.........)))..)))))))................. (-11.45 = -11.00 + -0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:55 2011