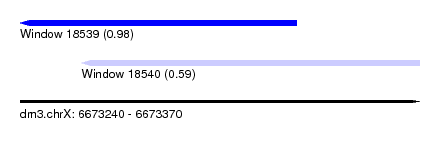
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,673,240 – 6,673,370 |
| Length | 130 |
| Max. P | 0.979874 |

| Location | 6,673,240 – 6,673,330 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.44010 |
| G+C content | 0.40511 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
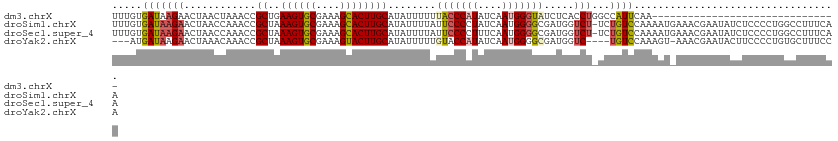
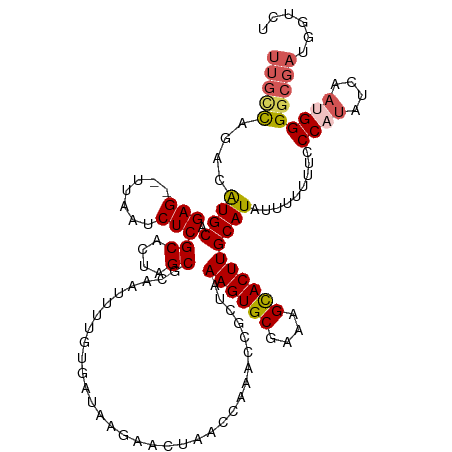
>dm3.chrX 6673240 90 - 22422827 UUUGUGAUAAGAACUAACUAAACCGCUGAAGUGCGAAAGCACUUGCAUAUUUUUUACCCAUAUCAAUGGGUAUCUCACCUGGCCAUUCAA------------------------------- ...((((.................((..((((((....))))))))........(((((((....)))))))..))))............------------------------------- ( -24.00, z-score = -3.27, R) >droSim1.chrX 5244965 120 - 17042790 UUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCCUAUCAAUGGGGCGAUGGUCU-UCUGUCCAAAAUGAAACGAAUAUCUCCCCUGGCCUUUCAA ((((.((((.(((...((((....((..((((((....)))))))).........(((((((....))))).)))))).)-)))))))))).......(((..((......))...))).. ( -29.00, z-score = -2.20, R) >droSec1.super_4 6057126 120 + 6179234 UUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCCUUUCAAUGGGGCGAUGGUCU-UCUGUCCAAAAUGAAACGAAUAUCUCCCCUGGCCUUUCAA ((((.((((.(((...((((....((..((((((....))))))))..........((((.......))))...)))).)-)))))))))).......(((..((......))...))).. ( -28.90, z-score = -2.06, R) >droYak2.chrX 2637509 113 - 21770863 ---AUGAUAAGAACUAAACAAACCGCUAAAGUGCGAAAGUACUUGCAUAUUUUUGUACCAUAUCAAUGGGGCGAUGGUC----UGUCCAAAGU-AAACGAAUACUUCCCCUGUGCUUUCCA ---.((..(((.((...(((.(((((..((((((....))))))))......(((..((((....))))..))).))).----)))...((((-(......))))).....)).)))..)) ( -24.70, z-score = -1.16, R) >consensus UUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCAUAUCAAUGGGGCGAUGGUCU_UCUGUCCAAAAU_AAACGAAUAUCUCCCCUGGCCUUUCAA .....((((...............((..((((((....))))))))........(((((((....)))))))...........)))).................................. (-16.49 = -16.93 + 0.44)
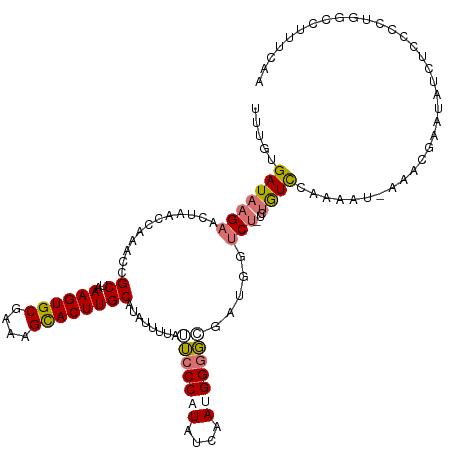
| Location | 6,673,260 – 6,673,370 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.26412 |
| G+C content | 0.43532 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6673260 110 - 22422827 UUGCUAGACAUGCAGAGUUUUAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACUAAACCGCUGAAGUGCGAAAGCACUUGCAUAUUUUUUACCCAUAUCAAUGGG---------- ..(((((...(((.(((.......)))))))))))((......(((.................)))..((((((....))))))))..........(((((....)))))---------- ( -29.23, z-score = -2.68, R) >droSim1.chrX 5245005 118 - 17042790 UUGCCAGGCGUGCAGAG--UUAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCCUAUCAAUGGGGCGAUGGUCU ..((((.((((((.(((--.....)))))))(((((....((((.(.((.....)).))))).)))))((((((....)))))))).........(((((((....))))).)))))).. ( -35.20, z-score = -1.94, R) >droSec1.super_4 6057166 117 + 6179234 UUGCCAGGCGUGCAGAG--UUAAUCUCGCGCUAGCGC-AUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCCUUUCAAUGGGGCGAUGGUCU .((((.((((((.(((.--....))))))))).).))-).................((((....((..((((((....))))))))..........((((.......))))...)))).. ( -37.00, z-score = -2.04, R) >droYak2.chrX 2637545 112 - 21770863 UUGCCAGACAUGCAGAG--UUAAUCUCGCAC-AGCGCAA-----UGAUAAGAACUAAACAAACCGCUAAAGUGCGAAAGUACUUGCAUAUUUUUGUACCAUAUCAAUGGGGCGAUGGUCU .....((((.(((.(((--.....))))))(-(.(((..-----.............(((((......((((((....)))))).......))))).((((....)))).))).)))))) ( -28.62, z-score = -1.34, R) >droEre2.scaffold_4690 15601551 112 - 18748788 UUGUCAGACAUGCAGAG--UUAAUCUCGCACUAGCACAA-----UGAUAAGAACUAACCCAACCGCCAAAGUGCGAAAGCACUUGCAUAUUUGU-UCCCACAUCAAUGGGGCGAUGGUGU ((((((........(((--.....)))((....))....-----)))))).............(((((((((((....))))))......((((-.((((......))))))))))))). ( -31.30, z-score = -1.75, R) >consensus UUGCCAGACAUGCAGAG__UUAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUUUUCCCAUAUCAAUGGGGCGAUGGUCU (((((....((((.(((.......)))((....)).................................((((((....)))))))))).........((((....)))))))))...... (-21.10 = -21.82 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:53 2011