
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,664,857 – 6,664,955 |
| Length | 98 |
| Max. P | 0.663157 |

| Location | 6,664,857 – 6,664,955 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 52.49 |
| Shannon entropy | 0.76014 |
| G+C content | 0.46426 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -10.21 |
| Energy contribution | -9.28 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
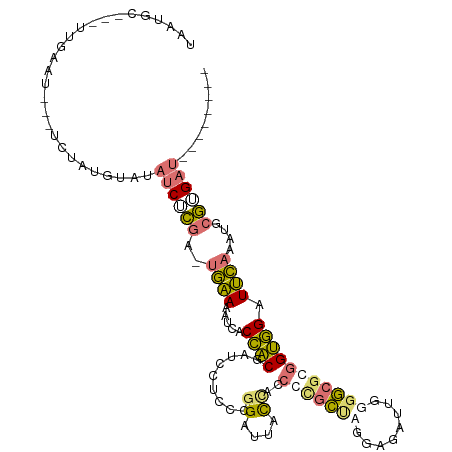
>dm3.chrX 6664857 98 + 22422827 --------AUCACGCAUUUGAAUCCACCGCGCCACAAUCUCCUAGCGGG-UAGUUACCCGGGACGAUUGUGGUAAUUUUCA-UUGAGAUACACAUAGA---AUCC-----GCAUUU --------....................((((((((((((((...((((-(....)))))))).)))))))))........-..(.(((.(.....).---))))-----)).... ( -26.30, z-score = -1.90, R) >droEre2.scaffold_4690 15593060 114 + 18748788 AGUGACGAGUCUCGCACUUGAAUCCACCCCGCCCCAAUCUCCUAGCGGGGAGGAAAUCCGG-AGGAUCGUGGUGAAGUUCA-UCGAGAUAUGUAUAUAUACAUUGAAUACGCAUUA ((((.((.((((((....(((((.((((.((((((..(((((....)))))((....))))-.))..)).))))..)))))-.))))))(((((....)))))......)))))). ( -33.70, z-score = -1.02, R) >droYak2.chrX 2629199 100 + 21770863 --------AUCACGCAUUGGAAACCACCCGGCGC-AAUCCCCCAGCGGGGCGGCAGGCCGGGAGGAUCGUGGUGAAAUUCA-UCGAGAUACACGUAGA---AUUUAA---GCAUUA --------.....((.(((((..((.((((((((-...((((....))))..))..)))))).))(((.(((((.....))-))).))).........---.)))))---)).... ( -34.10, z-score = -1.40, R) >droAna3.scaffold_13117 4449852 104 + 5790199 --------AGCAAGUAUUUAUACCAUCUGCGGCAGAUGCUUCUGCAAGGGUUAUCAUCUACUUAUAUAGAUGUCAUCUAUAGCCUUGUUGUAAACAAACAUAGUCUUGAUGA---- --------...............((((.((.(((((....))))).((((((((((((((......))))))......))))))))................))...)))).---- ( -22.10, z-score = -0.19, R) >consensus ________AUCACGCAUUUGAAUCCACCCCGCCACAAUCUCCUAGCGGGGUGGUAAUCCGGGAGGAUCGUGGUGAAAUUCA_UCGAGAUACACAUAGA___AUUCAA___GCAUUA ........((((((....((((.((((..........(((((....)))))....((((....)))).)))).....))))..))))))........................... (-10.21 = -9.28 + -0.94)


| Location | 6,664,857 – 6,664,955 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 52.49 |
| Shannon entropy | 0.76014 |
| G+C content | 0.46426 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -10.43 |
| Energy contribution | -12.43 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6664857 98 - 22422827 AAAUGC-----GGAU---UCUAUGUGUAUCUCAA-UGAAAAUUACCACAAUCGUCCCGGGUAACUA-CCCGCUAGGAGAUUGUGGCGCGGUGGAUUCAAAUGCGUGAU-------- ..((((-----.((.---((((((((......((-(....))).((((((((.((((((((....)-))))...)))))))))))))).))))).))....))))...-------- ( -32.00, z-score = -2.35, R) >droEre2.scaffold_4690 15593060 114 - 18748788 UAAUGCGUAUUCAAUGUAUAUAUACAUAUCUCGA-UGAACUUCACCACGAUCCU-CCGGAUUUCCUCCCCGCUAGGAGAUUGGGGCGGGGUGGAUUCAAGUGCGAGACUCGUCACU ..((((((.....)))))).............((-(((..(((..((((((((.-..)))))(((.(((((((..........))))))).))).....))).)))..)))))... ( -32.80, z-score = -0.46, R) >droYak2.chrX 2629199 100 - 21770863 UAAUGC---UUAAAU---UCUACGUGUAUCUCGA-UGAAUUUCACCACGAUCCUCCCGGCCUGCCGCCCCGCUGGGGGAUU-GCGCCGGGUGGUUUCCAAUGCGUGAU-------- ......---......---..(((((((...(((.-((........))))).((.((((((..((..((((....))))...-)))))))).))......)))))))..-------- ( -32.20, z-score = -0.74, R) >droAna3.scaffold_13117 4449852 104 - 5790199 ----UCAUCAAGACUAUGUUUGUUUACAACAAGGCUAUAGAUGACAUCUAUAUAAGUAGAUGAUAACCCUUGCAGAAGCAUCUGCCGCAGAUGGUAUAAAUACUUGCU-------- ----(((((.((.((.((((.(....))))).))))...)))))(((((((....))))))).........(((((....))))).(((...((((....))))))).-------- ( -22.00, z-score = -0.77, R) >consensus UAAUGC___UUGAAU___UCUAUGUAUAUCUCGA_UGAAAAUCACCACGAUCCUCCCGGAUUACCACCCCGCUAGGAGAUUGGGGCGCGGUGGAUUCAAAUGCGUGAU________ ............................(((((..((((.........(((((....))))).(((((((((((........))))))))))).))))....)))))......... (-10.43 = -12.43 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:50 2011