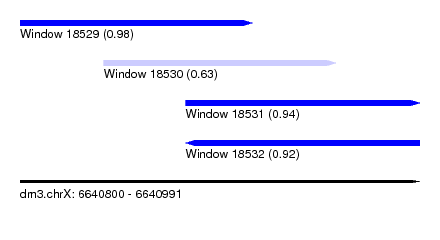
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,640,800 – 6,640,991 |
| Length | 191 |
| Max. P | 0.981764 |

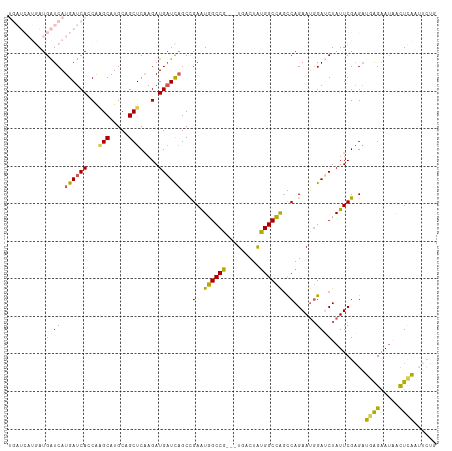
| Location | 6,640,800 – 6,640,911 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.37538 |
| G+C content | 0.53780 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6640800 111 + 22422827 UCACAAAAAAAAAAAAAACGAGGCGGGAUCAAGGGUAGCGUGAUCAUGAUGAUCAUGAUCAUCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAG .....................(((..(((((.((((.(((((....(((((((....)))))))..))))).))))....))))).)))...((((((---(....))))))). ( -39.80, z-score = -4.57, R) >droEre2.scaffold_4690 15566930 91 + 18748788 --------------UCAUAGAGGCGGGAUCAAGGGCAGC---------AUGGUCAUGAUCAGCGAGCAUGCAGCUCAAGAUGCUUAGCCGAGUGGCCGAGGUGACUGCGGCUGG --------------.......(((((.(((..((((.((---------(((.((.........)).))))).))))..))).))..)))....(((((((....)).))))).. ( -35.80, z-score = -1.59, R) >droYak2.chrX 2603905 88 + 21770863 --------------UCAUAGAGGCGGGAUCAAGGGCAGC---------AUGAUCAUGAUCAGCACGCAUGCAGCUCGAGAUGAUCAGCUGAGUGGCCG---UGACUGUGGCCAG --------------((((((..((((..(((....((((---------.(((((((..(((((.........))).)).)))))))))))..))))))---)..)))))).... ( -29.70, z-score = -0.14, R) >droSec1.super_4 6025782 103 - 6179234 --------UCACAAAAACCGAGGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCCGCCGAAUGGCCG---UGACUAUGGCCAA --------.............(((((.((((((.((((((((((((((.....)))))))...)))).)))..)).....)))))))))...((((((---(....))))))). ( -41.50, z-score = -3.16, R) >droSim1.chrX_random 2112183 103 + 5698898 --------UCACAAAAACCGAGGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAG --------.............(((..(((((((.((((((((((((((.....)))))))...)))).)))..)).....))))).)))...((((((---(....))))))). ( -39.20, z-score = -2.72, R) >consensus __________A_AAAAACCGAGGCGGGAUCAAGGGCAGC_UGAUCAUGAUGAUCAUGAUCACCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG___UGACUAUGGCCAG .....................(((..(((((.((((.((.((.......((((....)))).....)).)).))))....))))).)))...((((((.........)))))). (-24.32 = -24.20 + -0.12)

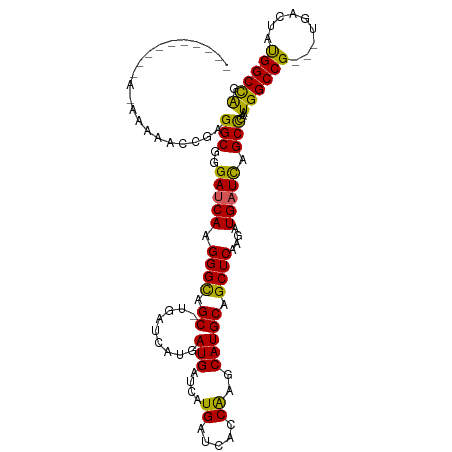
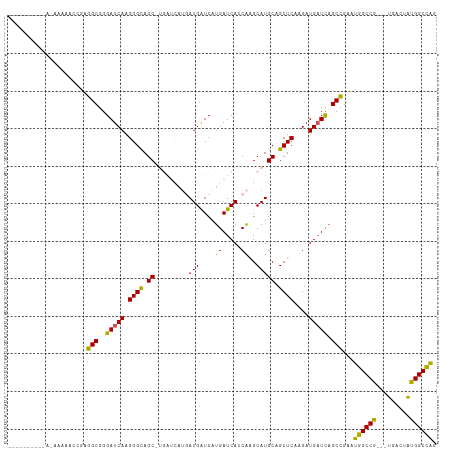
| Location | 6,640,840 – 6,640,951 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.55 |
| Shannon entropy | 0.38142 |
| G+C content | 0.48098 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -17.48 |
| Energy contribution | -16.84 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6640840 111 + 22422827 UGAUCAUGAUGAUCAUGAUCAUCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAGUUCAAUUCUG (((((.....)))))((((((((.(((.....)))...))))))))......((((((---(....)))))))..((((((.((.((((((.......)))))).)).)))))) ( -39.90, z-score = -2.87, R) >droEre2.scaffold_4690 15566955 101 + 18748788 --------AUGGUCAUGAUCAGCGAGCAUGCAGCUCAAGAUGCUUAGCCGAGUGGCCGAGGUGACUGCGGCUGGCCAGAACUGAGCUAUUCGCGAAAGGAACUACUUUG----- --------.((((....))))(((((.....((((((...((.((((((((((.((....)).))).))))))).))....)))))).))))).((((......)))).----- ( -33.90, z-score = -0.54, R) >droYak2.chrX 2603930 98 + 21770863 --------AUGAUCAUGAUCAGCACGCAUGCAGCUCGAGAUGAUCAGCUGAGUGGCCG---UGACUGUGGCCAGUCAGAAUGGAACUAUUCGAGCAAAGGAUCACUGUA----- --------.......(((((.(((....))).(((((((..(.((..((((.((((((---......)))))).))))....)).)..)))))))....))))).....----- ( -36.30, z-score = -2.03, R) >droSec1.super_4 6025814 111 - 6179234 UGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCCGCCGAAUGGCCG---UGACUAUGGCCAACCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAUUUCUG ((((((((.....))))))))...(((.....)))...((.((((((.....((((((---(....))))))).......))))))...))((((((((.....)))))))).. ( -37.40, z-score = -2.77, R) >droSim1.chrX_random 2112215 111 + 5698898 UGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAUUUCUG ((((((((.....))))))))...(((..((.(.((.....)).).))....((((((---(....)))))))))).(((((....)))))((((((((.....)))))))).. ( -38.20, z-score = -2.43, R) >consensus UGAUCAUGAUGAUCAUGAUCACCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG___UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAAUUCUG ..........((((.((((((...(((.....))).....))))))...(..((((((.........))))))..)......)))).........((((.....))))...... (-17.48 = -16.84 + -0.64)

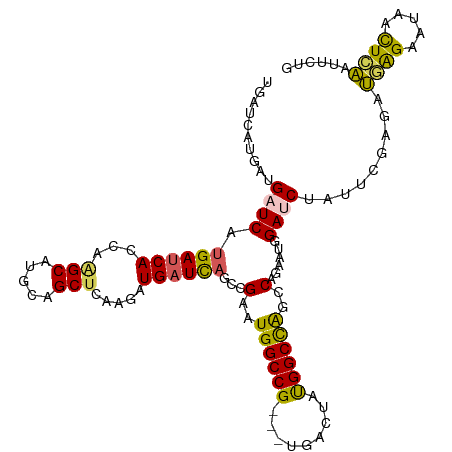
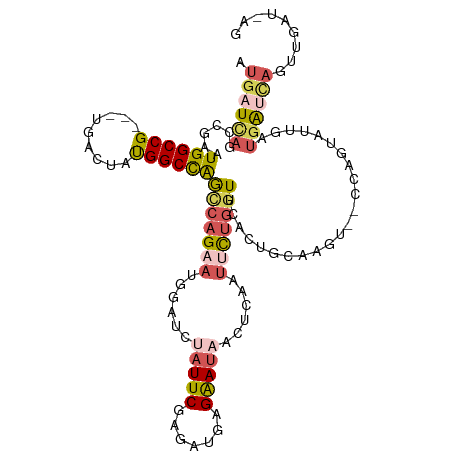
| Location | 6,640,879 – 6,640,991 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.53382 |
| G+C content | 0.46301 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -13.58 |
| Energy contribution | -15.02 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6640879 112 + 22422827 AUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAGUUCAAUUCUGUUUCACUGCAAGUCGCCAGUAUUGAUGAUCAGUUGAUUAG .((((((((.((.((((((---(....)))))))..((((((.((.((((((.......)))))).)).))))))....(((((.....).))))........)).)))))))). ( -35.60, z-score = -2.58, R) >droEre2.scaffold_4690 15566986 91 + 18748788 AUGCUUAGCCGAGUGGCCGAGGUGACUGCGGCUGGCCAGAACUGAGCUAUUCGCGAAAGGAACUACU-----UUGGUUCGGU--------CAAGUAUUGCUGAU----------- ((((((((((((((.((....)).))).)))))((((.((((..((.(((((.(....)))).)).)-----)..)))))))--------))))))).......----------- ( -34.30, z-score = -2.29, R) >droYak2.chrX 2603961 98 + 21770863 AUGAUCAGCUGAGUGGCCG---UGACUGUGGCCAGUCAGAAUGGAACUAUUCGAGCAAAGGAUCACUGUAUGUAAGUCUACUA-------UCGAUGAUGAUUGCUAGG------- .(((((.(((((.((((((---......)))))).)).(((((....))))).)))....)))))...............(((-------.((((....)))).))).------- ( -26.40, z-score = -0.40, R) >droSec1.super_4 6025853 110 - 6179234 AUGAUCCGCCGAAUGGCCG---UGACUAUGGCCAACCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAUUUCUGGUUCACUGCAAGU--CCAGUAUUGGUGAUCAGUUGAUAAG .(((((.(((((.((((((---(....)))))))(((((((((....)))))((((((((.....))))))))))))..((((.....--.)))).))))))))))......... ( -37.70, z-score = -3.31, R) >droSim1.chrX_random 2112254 110 + 5698898 AUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAUUUCUGGUUCACUGCAAGU--CCAGUAUUGGUGAUCAGUUGAUAAG .((((((.((((.((((((---(....)))))))(((((((((....)))))((((((((.....))))))))))))..((((.....--.)))).))))))))))......... ( -38.40, z-score = -3.22, R) >consensus AUGAUCAGCCGAAUGGCCG___UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAAUAACUCAAUUCUGGUUCACUGCAAGU__CCAGUAUUGAUGAUCAGUUGAU_AG .((((((......((((((.........))))))(((((((......(((((.......)))))......))))))).......................))))))......... (-13.58 = -15.02 + 1.44)

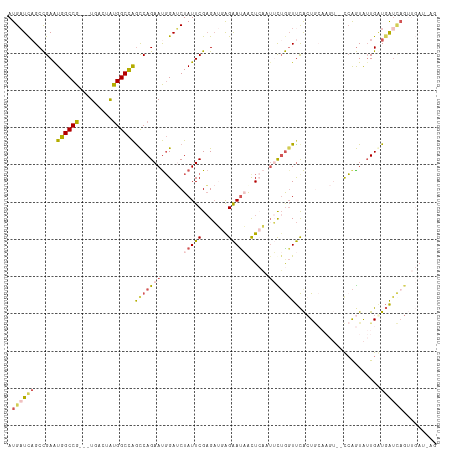
| Location | 6,640,879 – 6,640,991 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.53382 |
| G+C content | 0.46301 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6640879 112 - 22422827 CUAAUCAACUGAUCAUCAAUACUGGCGACUUGCAGUGAAACAGAAUUGAACUAUUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA---CGGCCAUUCGGCUGAUCAU .........((((((((...((((........))))(((.((((((.((.((((((.......)))))).)).))))))..(((((.......---.)))))))))).)))))). ( -32.50, z-score = -2.78, R) >droEre2.scaffold_4690 15566986 91 - 18748788 -----------AUCAGCAAUACUUG--------ACCGAACCAA-----AGUAGUUCCUUUCGCGAAUAGCUCAGUUCUGGCCAGCCGCAGUCACCUCGGCCACUCGGCUAAGCAU -----------....((.......(--------.((((((...-----(((.(((((....).)))).)))..)))).)).)(((((.(((..(....)..))))))))..)).. ( -21.20, z-score = -0.55, R) >droYak2.chrX 2603961 98 - 21770863 -------CCUAGCAAUCAUCAUCGA-------UAGUAGACUUACAUACAGUGAUCCUUUGCUCGAAUAGUUCCAUUCUGACUGGCCACAGUCA---CGGCCACUCAGCUGAUCAU -------..((((........((((-------(((..((.((((.....))))))..))).))))............(((.(((((.......---.))))).)))))))..... ( -21.70, z-score = -0.92, R) >droSec1.super_4 6025853 110 + 6179234 CUUAUCAACUGAUCACCAAUACUGG--ACUUGCAGUGAACCAGAAAUGAGUUAUUCUCAUCUCGAAUAGAUCCAUUCUGGUUGGCCAUAGUCA---CGGCCAUUCGGCGGAUCAU .........(((((.((..(((((.--.....)))))((((((((..((.((((((.......)))))).))..))))))))((((.......---.))))....))..))))). ( -32.80, z-score = -2.37, R) >droSim1.chrX_random 2112254 110 - 5698898 CUUAUCAACUGAUCACCAAUACUGG--ACUUGCAGUGAACCAGAAAUGAGUUAUUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA---CGGCCAUUCGGCUGAUCAU .........((((((((..(((((.--.....)))))..((((((..((.((((((.......)))))).))..)))))).(((((.......---.)))))...)).)))))). ( -32.70, z-score = -2.53, R) >consensus CU_AUCAACUGAUCACCAAUACUGG__ACUUGCAGUGAACCAGAAAUGAGUUAUUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA___CGGCCAUUCGGCUGAUCAU ....................................................................((((....((((.(((((...........))))).))))..)))).. (-13.46 = -13.70 + 0.24)


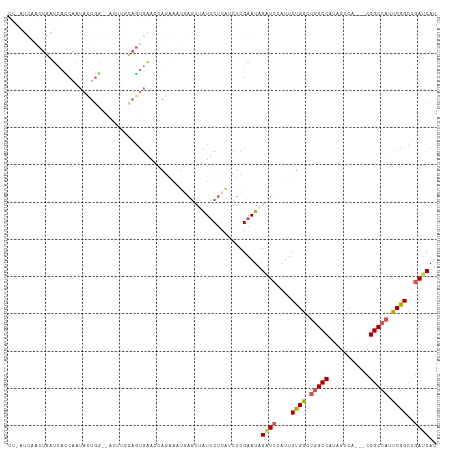
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:46 2011