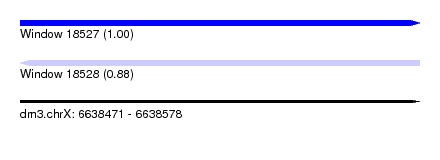
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,638,471 – 6,638,578 |
| Length | 107 |
| Max. P | 0.997590 |

| Location | 6,638,471 – 6,638,578 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Shannon entropy | 0.38671 |
| G+C content | 0.43352 |
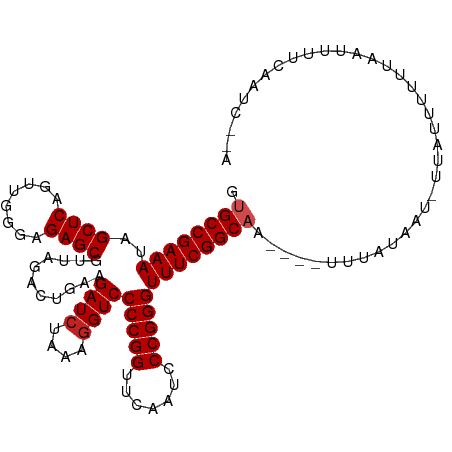
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -29.89 |
| Energy contribution | -29.91 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6638471 107 + 22422827 UUCGAUAGAAAAAUAAAAAAUAA-GCUAAGAA----UUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC (((....))).............-........----.(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.40, z-score = -2.88, R) >droVir3.scaffold_13042 2883520 105 - 5191987 ------UACUAAUUGAAAAAAAA-AAGAAAAAGGACUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGC ------.................-..............((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))).. ( -29.60, z-score = -2.39, R) >droSim1.chrX 5235135 107 + 17042790 UUAGAUUGAAAAUUAAAAAAUAA-GUUAUAAA----UUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC .......................-........----.(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.20, z-score = -3.12, R) >droSec1.super_4 6023455 107 - 6179234 UUAGUUUGAAAAUUAAAAAAUAA-GUUAUAAA----UUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC ((((((....)))))).......-........----.(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.30, z-score = -3.01, R) >droYak2.chrX 2601412 111 + 21770863 UUUGAUUGAAAAAUAAAAAAUAA-AAUAGAGAAUCAUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAU ..(((((................-.......))))).(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.40, z-score = -2.85, R) >droEre2.scaffold_4690 15564533 108 + 18748788 UUUGAUUGAAAAAUAAAAAAUAA-AACGGAGA---AUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC .......................-........---..(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.20, z-score = -2.78, R) >dp4.chrXL_group1a 6454786 109 + 9151740 UAUGAAAGAAAGCGAACAAAAAA-AUUAUUUUG--AAUGCCGAAACCCGGGAUCGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC ................(((((..-....)))))--..(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -31.60, z-score = -3.06, R) >droPer1.super_25 582765 105 - 1448063 ----UAUGAAAGCGAACCAAAAA-AGUGUUUUG--AAUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC ----..(.((((((.........-..)))))).--).(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -31.30, z-score = -2.56, R) >anoGam1.chr3L 28814133 99 - 41284009 ------AAAAAAUCAAAAAGUAG-AUUUAUUC----CUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGC-- ------...(((((........)-))))....----..((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))-- ( -29.60, z-score = -2.84, R) >droGri2.scaffold_15203 6859543 108 - 11997470 ----UGUCAAAAUAAUAAAUCGAUGUCGUCUUUGCUCUGCCGAAACCCGGGAUUGAACCGGGGACAUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAA ----.................................(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -30.40, z-score = -2.20, R) >droWil1.scaffold_180701 1777985 100 + 3904529 ------AAGGAGACAAAAAAUGA-AUUUGUAA-----UGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC ------..(....).........-........-----(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). ( -32.00, z-score = -3.15, R) >consensus U__GAUUGAAAAUUAAAAAAUAA_AUUAUAAA____UUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAC .....................................(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))). (-29.89 = -29.91 + 0.02)



| Location | 6,638,471 – 6,638,578 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Shannon entropy | 0.38671 |
| G+C content | 0.43352 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -26.37 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6638471 107 - 22422827 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAA----UUCUUAGC-UUAUUUUUUAUUUUUCUAUCGAA .((((((((..((((........))))...........((((....))))(((((.......))))))))))))).----........-....................... ( -27.60, z-score = -0.52, R) >droVir3.scaffold_13042 2883520 105 + 5191987 GCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAGUCCUUUUUCUU-UUUUUUUUCAAUUAGUA------ ..(((((((..((((........))))...........((((....))))(((((.......))))))))))))..............-.................------ ( -27.00, z-score = -0.48, R) >droSim1.chrX 5235135 107 - 17042790 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAA----UUUAUAAC-UUAUUUUUUAAUUUUCAAUCUAA .((((((((..((((........))))...........((((....))))(((((.......))))))))))))).----........-....................... ( -27.60, z-score = -1.32, R) >droSec1.super_4 6023455 107 + 6179234 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAA----UUUAUAAC-UUAUUUUUUAAUUUUCAAACUAA .((((((((..((((........))))...........((((....))))(((((.......))))))))))))).----........-....................... ( -27.60, z-score = -1.32, R) >droYak2.chrX 2601412 111 - 21770863 AUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAUGAUUCUCUAUU-UUAUUUUUUAUUUUUCAAUCAAA .((((((((..((((........))))...........((((....))))(((((.......))))))))))))).............-....................... ( -27.70, z-score = -0.95, R) >droEre2.scaffold_4690 15564533 108 - 18748788 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAU---UCUCCGUU-UUAUUUUUUAUUUUUCAAUCAAA .((((((((..((((........))))...........((((....))))(((((.......)))))))))))))..---........-....................... ( -27.60, z-score = -1.04, R) >dp4.chrXL_group1a 6454786 109 - 9151740 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCGAUCCCGGGUUUCGGCAUU--CAAAAUAAU-UUUUUUGUUCGCUUUCUUUCAUA ((((((((((.((((........))))((..(((((..((((....))))..)))))..)).....)))))))))).--(((((....-..)))))................ ( -29.90, z-score = -1.04, R) >droPer1.super_25 582765 105 + 1448063 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAUU--CAAAACACU-UUUUUGGUUCGCUUUCAUA---- (((((((((..((((........))))...........((((....))))(((((.......))))))))))))))(--(((((....-.))))))............---- ( -29.30, z-score = -0.88, R) >anoGam1.chr3L 28814133 99 + 41284009 --GCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAG----GAAUAAAU-CUACUUUUUGAUUUUUU------ --(((((((..((((........))))...........((((....))))(((((.......))))))))))))..----....((((-(........)))))...------ ( -28.20, z-score = -0.89, R) >droGri2.scaffold_15203 6859543 108 + 11997470 UUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAUGUCCCCGGUUCAAUCCCGGGUUUCGGCAGAGCAAAGACGACAUCGAUUUAUUAUUUUGACA---- ....(((((((((((.((((..(..(((((((((....).))).))))).(((((.......))))))..)))).)))).....((....))......)))))))...---- ( -28.10, z-score = -0.67, R) >droWil1.scaffold_180701 1777985 100 - 3904529 GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCA-----UUACAAAU-UCAUUUUUUGUCUCCUU------ (((((((((..((((........))))...........((((....))))(((((.......)))))))))))))-----).(((((.-......)))))......------ ( -29.20, z-score = -1.57, R) >consensus GUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAA____UUUAUAAU_UUAUUUUUUAAUUUUCAAUC__A .((((((((..((((........))))...........((((....))))(((((.......)))))))))))))..................................... (-26.37 = -26.65 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:43 2011