
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,632,529 – 6,632,604 |
| Length | 75 |
| Max. P | 0.898072 |

| Location | 6,632,529 – 6,632,604 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.46981 |
| G+C content | 0.47109 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -8.99 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6632529 75 + 22422827 UAUUGCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGUUCCCGA-------UACCCGUUGCAACUGGUUAUACCGGUUCGUCU ....................((...((((((((..(....)..-------))))))))))((((((.....))))))..... ( -17.70, z-score = -1.22, R) >droSim1.chrX 5224333 68 + 17042790 UAUUGCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGUUC--------------CCGUUGCAACCGGUUAUACCGGUUCGUCU ...(((((.......(((((.(((....))))))))...--------------...)))))(((((.....)))))...... ( -19.62, z-score = -2.66, R) >droSec1.super_4 6017385 75 - 6179234 UAUUGCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGCUCCCGA-------UACCCGUUGCAACCGGUUAUACCGGUUCGUCU ....................((...((((((((..(....)..-------))))))))))((((((.....))))))..... ( -19.60, z-score = -1.71, R) >droYak2.chrX 2595210 75 + 21770863 UAUUGCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGUUCCCAA-------UACCCGUUGCAACCGGUUAUACCGGUUCAUCU ....................((...((((((((..........-------))))))))))((((((.....))))))..... ( -19.30, z-score = -2.67, R) >droEre2.scaffold_4690 15558397 82 + 18748788 UAUUUCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGUUUCCAACCGGUUAUACCGGUUGCAACCGGCUAUACCGCUUCAUCU ........................((((((.((.((((..(((((((.....))))))).....)))).))))))))..... ( -21.20, z-score = -2.48, R) >dp4.chrXL_group1a 6447589 68 + 9151740 --UGGCAGACCUCUGUCGGAGCCAUUUCCAAUACAGUGUUC------------CCGUUGCAACCGGUUAAACCGGUUGUACU --.(((((....)))))(((((.(((........)))))))------------)...(((((((((.....))))))))).. ( -25.30, z-score = -3.44, R) >droPer1.super_25 575371 68 - 1448063 --UGGCAGACCUCUGUCGAAGCCAUUUCCAAUACAGUGUUC------------CCGUUGCAACCGGUUAAACCGGUUGGACU --((((.(((....)))...)))).................------------..(((.(((((((.....)))))))))). ( -20.30, z-score = -1.59, R) >consensus UAUUGCAAAAUUUUCCUUUAGCCCAAGCGGGUAAAGUUCCC_A_______UACCCGUUGCAACCGGUUAUACCGGUUCGUCU ....((((.......(((((.(((....))))))))....................))))((((((.....))))))..... ( -8.99 = -9.77 + 0.78)

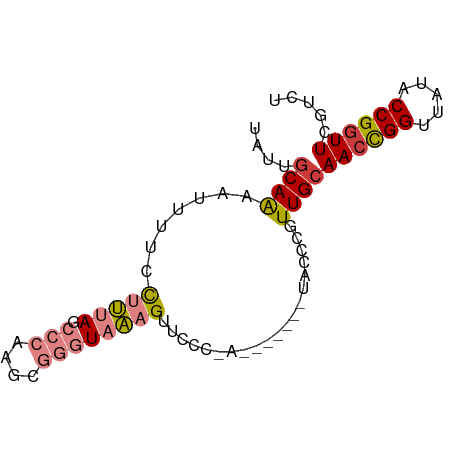
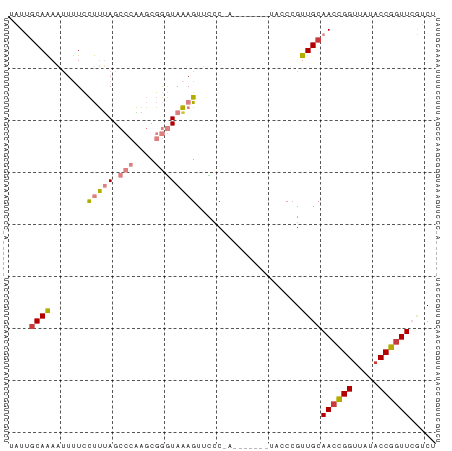
| Location | 6,632,529 – 6,632,604 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.46981 |
| G+C content | 0.47109 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -11.58 |
| Energy contribution | -11.01 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6632529 75 - 22422827 AGACGAACCGGUAUAACCAGUUGCAACGGGUA-------UCGGGAACUUUACCCGCUUGGGCUAAAGGAAAAUUUUGCAAUA .........((.....)).(((((((((((((-------..(....)..))))))(((......))).......))))))). ( -19.70, z-score = -0.99, R) >droSim1.chrX 5224333 68 - 17042790 AGACGAACCGGUAUAACCGGUUGCAACGG--------------GAACUUUACCCGCUUGGGCUAAAGGAAAAUUUUGCAAUA .....((((((.....))))))((((...--------------...((((((((....))).))))).......)))).... ( -19.92, z-score = -1.93, R) >droSec1.super_4 6017385 75 + 6179234 AGACGAACCGGUAUAACCGGUUGCAACGGGUA-------UCGGGAGCUUUACCCGCUUGGGCUAAAGGAAAAUUUUGCAAUA .....((((((.....))))))((((((((((-------..(....)..))))))(((......))).......)))).... ( -22.90, z-score = -1.43, R) >droYak2.chrX 2595210 75 - 21770863 AGAUGAACCGGUAUAACCGGUUGCAACGGGUA-------UUGGGAACUUUACCCGCUUGGGCUAAAGGAAAAUUUUGCAAUA .....((((((.....))))))((((((((((-------..(....)..))))))(((......))).......)))).... ( -24.80, z-score = -2.77, R) >droEre2.scaffold_4690 15558397 82 - 18748788 AGAUGAAGCGGUAUAGCCGGUUGCAACCGGUAUAACCGGUUGGAAACUUUACCCGCUUGGGCUAAAGGAAAAUUUUGAAAUA .....((.(((.....))).)).(((((((.....)))))))....((((((((....))).)))))............... ( -24.30, z-score = -1.67, R) >dp4.chrXL_group1a 6447589 68 - 9151740 AGUACAACCGGUUUAACCGGUUGCAACGG------------GAACACUGUAUUGGAAAUGGCUCCGACAGAGGUCUGCCA-- ....(((((((.....)))))))..((((------------.....))))........((((.((......))...))))-- ( -21.80, z-score = -1.32, R) >droPer1.super_25 575371 68 + 1448063 AGUCCAACCGGUUUAACCGGUUGCAACGG------------GAACACUGUAUUGGAAAUGGCUUCGACAGAGGUCUGCCA-- ....(((((((.....)))))))..((((------------.....))))..(((....((((((....))))))..)))-- ( -23.80, z-score = -1.91, R) >consensus AGACGAACCGGUAUAACCGGUUGCAACGGGUA_______U_GGGAACUUUACCCGCUUGGGCUAAAGGAAAAUUUUGCAAUA .....((((((.....))))))........................((((((((....))).)))))............... (-11.58 = -11.01 + -0.57)

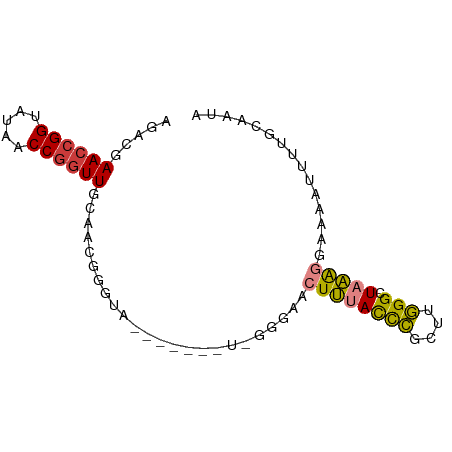
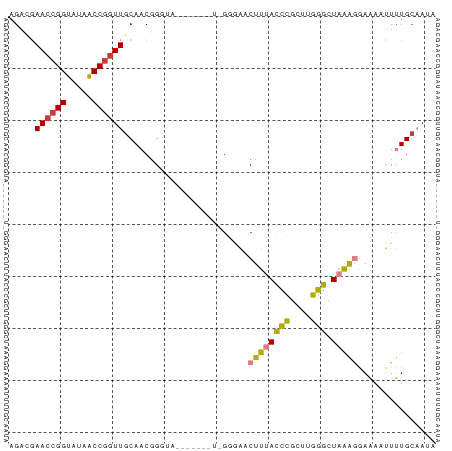
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:42 2011