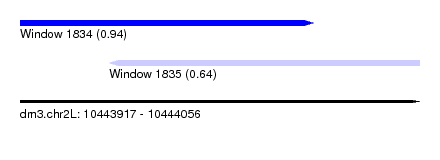
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,443,917 – 10,444,056 |
| Length | 139 |
| Max. P | 0.942905 |

| Location | 10,443,917 – 10,444,019 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Shannon entropy | 0.18203 |
| G+C content | 0.52113 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.942905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
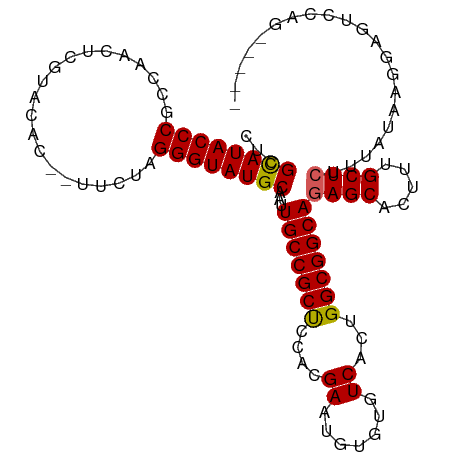
>dm3.chr2L 10443917 102 + 23011544 CUCGCAUACCCGCCAACUUGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAAUCCAG----- ...((((((((..............--.....))))))))...(((((((.((((....)))).....)))))))((((.....))))................----- ( -32.11, z-score = -2.10, R) >droEre2.scaffold_4929 11652976 106 - 26641161 CUCGCAUACCCGCUA-CUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCCCAAAGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUCAUAAGGAGUCUCCUCUGG ...((((((((....-.........--.....))))))))...(((((((....((.......))...)))))))((((.....)))).......((((....)))).. ( -34.27, z-score = -1.95, R) >droYak2.chr2L 6849022 108 + 22324452 CUCGUAUACCCGCUA-CUCACACACAGUUCUAGGGUAUGCAAUUGCCGCCCAACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUCUCCUCUGG .........((((((-.(.(((((((.(((..((((..((....)).))))...)))))))))).).))))))(((((......((((((....))))))...))))). ( -36.40, z-score = -2.55, R) >droSec1.super_3 5871819 102 + 7220098 CACGCAUACCCGCCAACUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAUAGCACUUUGCUCUUUAUAAGGAGUCCAG----- ...((((((((..............--.....))))))))...(((((((.((((....)))).....))))))).....((..((((((....))))))..))----- ( -30.91, z-score = -1.66, R) >droSim1.chr2L 10238036 102 + 22036055 CACGCAUACCCGCCAACUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUCCAG----- ...((((((((..............--.....))))))))...(((((((.((((....)))).....)))))))((((.....))))................----- ( -32.11, z-score = -1.70, R) >consensus CUCGCAUACCCGCCAACUCGUACAC__UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUCCAG_____ ...((((((((.....................))))))))...(((((((....((.......))...)))))))((((.....))))..................... (-29.36 = -29.16 + -0.20)

| Location | 10,443,948 – 10,444,056 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Shannon entropy | 0.35829 |
| G+C content | 0.45330 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
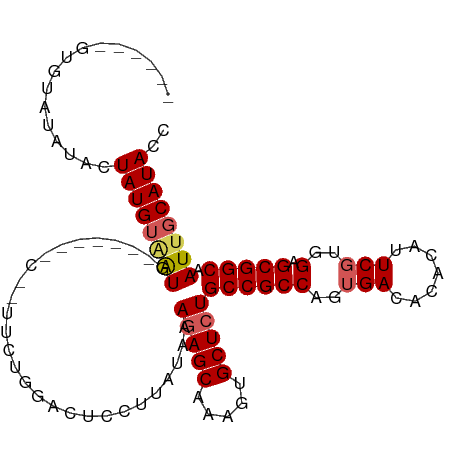
>dm3.chr2L 10443948 108 - 23011544 GUAUGUGUGUAUGUACUAUGUUAUGUUGUUAGCAUUUCUGGAUUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACC ((((((.(((.(((.(((((..(((((((((......((((...........(((((.....)))))....))))))))).)))).))))).))))))...)))))). ( -28.46, z-score = -0.24, R) >droEre2.scaffold_4929 11653006 99 + 26641161 --------GUAUAUA-UAUGGGGUUAGCAGUUCCAGAGGAGACUCCUUAUGAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCUUUGGGCGGCAAUUGCAUACC --------.......-.....(((..((((((...(((....))).......(((((.....)))))((((((...((.......))....))))))))))))..))) ( -31.90, z-score = -1.80, R) >droYak2.chr2L 6849054 103 - 22324452 ----GUAUGUACAUA-UAUGUAGUUAGCAGUUCCAGAGGAGACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUUGGGCGGCAAUUGCAUACC ----((((.((((..-..))))....((((((...(((....))).......(((((.....)))))((((((...(((........))).)))))))))))))))). ( -33.90, z-score = -2.87, R) >droSec1.super_3 5871850 92 - 7220098 ------GUAUAUAUACUAUGUAAUG----------UUCUGGACUCCUUAUAAAGAGCAAAGUGCUAUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACC ------..........(((((((((----------((((.............))))).........(((((((..(((.......)))..).)))))))))))))).. ( -22.32, z-score = -0.54, R) >droSim1.chr2L 10238067 92 - 22036055 ------GUAUAUAUACUAUGUAAUG----------UUCUGGACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACC ------..........((((((((.----------.................(((((.....)))))((((((..(((.......)))..).))))).)))))))).. ( -24.10, z-score = -1.11, R) >consensus ______GUGUAUAUACUAUGUAAUG_______C__UUCUGGACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACC ................((((((((............................(((((.....)))))((((((..(((.......)))..).))))).)))))))).. (-20.18 = -20.62 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:11 2011