
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,615,403 – 6,615,504 |
| Length | 101 |
| Max. P | 0.989721 |

| Location | 6,615,403 – 6,615,504 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.65044 |
| G+C content | 0.39875 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -12.71 |
| Energy contribution | -13.11 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
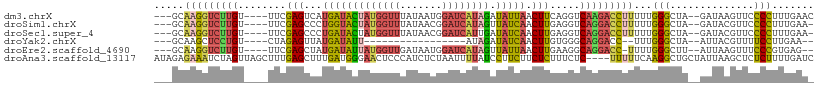
>dm3.chrX 6615403 101 - 22422827 ---GCAAGGUCUUGU----UUCGAGUCAUGAUACUAUGGUUUAUAAUGGAUCAUAGAUAUUAACUUCAGGUCAAGACCUUUUUGGGCUA--GAUAAGUUCCCCUUUGAAC ---..((((((((((----((.((((..((((((((((((((.....))))))))).))))))))).))).)))))))))...(((...--((.....)))))....... ( -27.60, z-score = -2.14, R) >droSim1.chrX 5206297 100 - 17042790 ---GCAAGGUCUUGU----UUCGAGCCCUGGUACUAUGGUUUAUAACGGAUCAUAGUUAUCAACUUGAGGUCAGGACCUUUUUGGGCUA--GAUACGUUCCCCUUUGAA- ---..((((((((((----((((((...((((((((((((((.....))))))))).))))).))))))).)))))))))...(((...--((.....)))))......- ( -32.40, z-score = -2.41, R) >droSec1.super_4 6000936 100 + 6179234 ---GCAAGGUCUUGU----UUCGAGCCCUGAUACUAUGGUUUAUAACGGAUCAUUGAUAUCAACUUGAGGUCAGGACCUUUUUGGGCUA--GAUACGUUCCCCUUUGAA- ---..((((((((((----((((((...((((((.(((((((.....))))))).).))))).))))))).)))))))))...(((...--((.....)))))......- ( -28.50, z-score = -1.62, R) >droYak2.chrX 2577465 80 - 21770863 ---GCAAGCUCCUGU----CUAGAGUUAUGAUAUU-----------------AUAGAUAUCAACUUGUGGGCAGGACC--UUUGGGCUA--AUUACGUUUUCCUUGAA-- ---.((((.((((((----(((((((..(((((((-----------------...))))))))))).)))))))))((--....))...--...........))))..-- ( -21.70, z-score = -1.72, R) >droEre2.scaffold_4690 15542172 98 - 18748788 ---GCAAGGUCUUGU----UUCGAGCUAUGAUAUUAUGGUUGAUAAUGGAUCAUAGUUAUUAACUUGAAGGCAGGACC-UUUUGGGCUU--AUUAAGUUUCCCGUGAG-- ---((((((((((((----((((((.((....(((((((((.......)))))))))...)).)))))).))))))))-))..(((...--.........)))))...-- ( -27.80, z-score = -2.27, R) >droAna3.scaffold_13117 4384345 106 - 5790199 AUAGAGAAAUCUAGUUAGCUUUGAGCUUUGAUGGGAACUCCCAUCUCUAAUUUUAUCCUUCUUCUCUUUCUC----UUUUUCAAGGCUGCUAUUAAGCUCUCUUUUGAUC ..((((.....((((.(((((((((....((((((....))))))..((....)).................----...))))))))))))).....))))......... ( -23.30, z-score = -1.21, R) >consensus ___GCAAGGUCUUGU____UUCGAGCCAUGAUACUAUGGUUUAUAACGGAUCAUAGAUAUCAACUUGAGGUCAGGACCUUUUUGGGCUA__AAUAAGUUCCCCUUUGAA_ .....(((((((((........(((...(((((((((((((.......)))))))).))))).))).....)))))))))...(((..............)))....... (-12.71 = -13.11 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:35 2011