| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,604,327 – 6,604,426 |
| Length | 99 |
| Max. P | 0.737113 |

| Location | 6,604,327 – 6,604,426 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.40100 |
| G+C content | 0.32243 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -11.23 |
| Energy contribution | -12.71 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
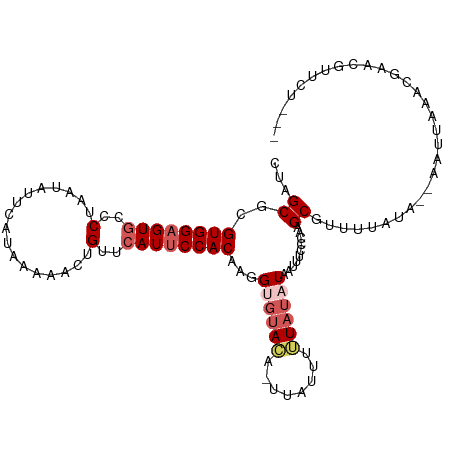
>dm3.chrX 6604327 99 + 22422827 CUAGCGCGUGGAGUGCCCUAAUAUUCAUAAAAACUGUUCAUUCCACAAGGUGUACACUUAUUUUUAUAUAAUUUCCGAAGCGUUUUAUA--AGUUUUUUUU---------- ..(((((((((((((..(.................)..)))))))).((((....))))....................))))).....--..........---------- ( -18.03, z-score = -1.55, R) >droEre2.scaffold_4690 15531540 100 + 18748788 CUAGCACGUGGUGUAGACUAAUAUUCAUAAAAA-CGUUCAUUCCACAAGGGGUAA---U-UUUUUAUAUAAUUCCUCAAGCAUUUUUA---AAUUCAACGAAAGUUCU--- ...((..((((.((..((...............-.))..)).))))..((((.((---(-(........))))))))..)).......---.....(((....)))..--- ( -17.19, z-score = -2.02, R) >droYak2.chrX 2567110 107 + 21770863 CUAGCACGUGGAGUGGCCUAAUAUUCAUAAAAA-UGUUCAUUCCACAAUGGGAAA---CAUUUUUAUAUAAUUUAUCAAGCAUUUUUUUUUAUUUCAAUGAAUGUUCUAAU .(((.((((.(((((......)))))(((((((-((((..((((.....))))))---)))))))))..................................)))).))).. ( -17.50, z-score = -0.91, R) >droSec1.super_4 5989952 105 - 6179234 CUAGCGCGUGGAGUGCCCUAAUAUUCAUAAAAACUGUUCAUUCCACAAGGUGUACAUUUAUUUGUAGAUUAUUUCCCAAGCGUUUUACA--AAUCAAACGAACGGUU---- ..(((((((((((((..(.................)..))))))))..(((.((((......)))).))).........))))).....--................---- ( -21.13, z-score = -1.26, R) >droSim1.chrX 5194990 107 + 17042790 CUAGCGCGUGGAGUGCCCUAAUAUUCAUAAAAACUGUUCAUUCCACAAGGUGUACAUUUAUUUGUAUAUUAUUUCCCAAGCGUUUUAUA--AAUCAAACGAACGUUUUU-- ..(((((((((((((..(.................)..))))))))..((((((((......)))))))).........))))).....--....((((....))))..-- ( -23.43, z-score = -2.82, R) >consensus CUAGCGCGUGGAGUGCCCUAAUAUUCAUAAAAACUGUUCAUUCCACAAGGUGUACA_UUAUUUUUAUAUAAUUUCCCAAGCGUUUUAUA__AAUUAAACGAACGUUCU___ ...((..((((((((..(.................)..))))))))...(((((((......)))))))..........)).............................. (-11.23 = -12.71 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:34 2011