| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,587,942 – 6,588,033 |
| Length | 91 |
| Max. P | 0.843951 |

| Location | 6,587,942 – 6,588,033 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Shannon entropy | 0.32682 |
| G+C content | 0.44136 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6587942 91 - 22422827 UAGACGACAAUUUCGAUUGUUGUUG-CCCCGGCUUUUGACCAAGAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCUCACUUGAUCA------- ..(((((((((....))))))))).-....((((((...(((....)))......(((((...)))))......))))))............------- ( -21.60, z-score = -1.51, R) >droSim1.chrX_random 2100209 92 - 5698898 UAGACGACAAUUUAGAUGGUUGUAGCCCCCGGCUUGAGACCAAGAAAGGAACCAGAUCCCAAAGGGAUAAAAAUAGAGCCCUCAGUUGAUCA------- ..((((((........(((((..((((...))))...))))).((..((......(((((...)))))..........)).)).)))).)).------- ( -20.99, z-score = -1.41, R) >droSec1.super_4 5973731 92 + 6179234 UAGACGACAAUUUCGAUUGUUGUUGCCCCCGGCUUUUGACCAAGAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCUCACUUGAUCA------- ..(((((((((....)))))))))......((((((...(((....)))......(((((...)))))......))))))............------- ( -21.60, z-score = -1.42, R) >droYak2.chrX 2551135 92 - 21770863 UAGACGACAAUUUCGAUUGUUGUUGCCCCCGGCUUUUGACCAAGAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCUCACUUGAUCA------- ..(((((((((....)))))))))......((((((...(((....)))......(((((...)))))......))))))............------- ( -21.60, z-score = -1.42, R) >droEre2.scaffold_4690 15514925 92 - 18748788 UAGACGACAAUUUCGAUUGUUGUUGCCCCCGGCUUUUGACCAAGAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCUCACUUGAUCA------- ..(((((((((....)))))))))......((((((...(((....)))......(((((...)))))......))))))............------- ( -21.60, z-score = -1.42, R) >droAna3.scaffold_13117 4344192 93 - 5790199 -UGACGACAAUUUCGAUUGUUGU----UCAGGCUUU-AGCGAAUAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCACACUUGACCACCACCAA -.(((((((((....))))))))----)..((((((-(........((....)).(((((...))))).....)))))))................... ( -21.20, z-score = -1.95, R) >dp4.chrXL_group1a 394143 94 - 9151740 GAGACGACAAUUUCGAUUGUUGU----UCCGGCUU-UGACCAAUAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCACACUUGACCAUCACCAA (..((((((((....))))))))----..)(((((-((.(((....)))......(((((...))))).....)))))))................... ( -24.30, z-score = -2.68, R) >droPer1.super_26 1077924 94 + 1349181 GAGACGACAAUUUCGAUUGUUGU----UCCGGCUU-UGACCAAUAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCACACUUGACCAUCACCAA (..((((((((....))))))))----..)(((((-((.(((....)))......(((((...))))).....)))))))................... ( -24.30, z-score = -2.68, R) >droWil1.scaffold_181096 9872642 93 - 12416693 --GACGACAACUUUGAUUGUUGU----UCAGGCCCUGGCCAAAUAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCGACACUUGACCACCAUCAG --((((((((......)))))))----)..(((.(((((((.....)))..))))(((((...))))).........)))................... ( -21.90, z-score = -1.53, R) >droMoj3.scaffold_6359 4434428 91 - 4525533 --GACGACAAUUUCGAUUGUUGU----UCAGGCUUU-GCCAAAUAAUGGAACCAGAUCCCAAUGGGAUAAAGAUCGAACCAACACUUGACCAUCACCA- --..........((((.((((((----((.(((...-)))........((.(...(((((...)))))...).))))).))))).)))).........- ( -18.70, z-score = -0.73, R) >droGri2.scaffold_14853 5047980 91 - 10151454 --GACGACAAUUUCGAUUGUUGU----UCAGGCUUU-GCCAAAUAAUGGAACCAGAUCCCAAUGGGAUAAAGAUAGAGCCGACACUUGACCAUCAUCA- --(((((((((....))))))))----)..((((((-((((.....)))......(((((...))))).....)))))))..................- ( -22.70, z-score = -1.82, R) >consensus UAGACGACAAUUUCGAUUGUUGU____UCCGGCUUUUGACCAAUAAUGGAACCAGAUCCCAAUGGGAUAAAAAUAGAGCCCACACUUGACCA_CA_CA_ ...((((((((....)))))))).......(((((...........((....)).(((((...))))).......)))))................... (-16.09 = -16.46 + 0.37)

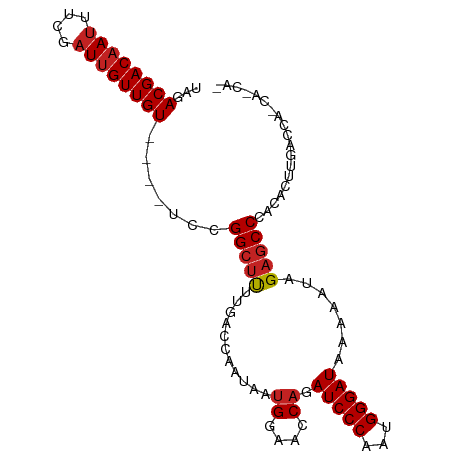

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:31 2011