
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,586,019 – 6,586,155 |
| Length | 136 |
| Max. P | 0.996667 |

| Location | 6,586,019 – 6,586,127 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 97.59 |
| Shannon entropy | 0.04034 |
| G+C content | 0.34444 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -23.61 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6586019 108 + 22422827 GCAUUUUCCUUUGAGGCAGUUGAUUUCGCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAGUAUUGGCAAUUGUUUGAUAUUUUACCAGCGCUACGAAUUUGUAUUCG (((((((((((....(((((((.......)))))))))))))))).............((((((..((....).)..)))))).....))....(((((....))))) ( -25.60, z-score = -1.78, R) >droSec1.super_4 5971733 108 - 6179234 UCAUUUUCCUUUGAGGCAGUUGAUUUCGCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAAUAUUGGCAAUUGUUUGAUAUUUUACCAGCGCUACGAAUUUGUAUUCG ..(((((((((....(((((((.......))))))))))))))))...((((((...((((((........))))))..)))))).........(((((....))))) ( -24.40, z-score = -1.83, R) >droYak2.chrX 2549172 108 + 21770863 UCAUUUUCCUUGGAGGCAGUUGAUUUCUCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAGUAUUGGCAAUUGUUUGAUAUUUUACCAGUGCUACGAAUUUGUAUUCG ..(((((((((....(((((((.......)))))))))))))))).....((((((...((((((((.((............)).))))))))..))))))....... ( -27.60, z-score = -2.66, R) >droEre2.scaffold_4690 15512937 108 + 18748788 UCAUUUUCCUUUGAGGCAGUUGAUUUCUCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAAUAUUGGCAAUUGUUUGAUAUUUUACCAGUGCUACGAAUUUGUAUUCG ..(((((((((....(((((((.......))))))))))))))))....(((...(((((((..(((((.(((..(((....))).))))))))...)))))))))). ( -26.30, z-score = -2.81, R) >droSim1.chrX_random 2097937 108 + 5698898 UCAUUUUCCUUUGAGGCAGUUGAUUUCGCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAGUAUUGGCAAUUGUUUGAUAUUUUACCAGCGCUACGAAUUUGUAUUCG ..(((((((((....(((((((.......)))))))))))))))).....((((((...((..((((.((............)).))))..))..))))))....... ( -24.00, z-score = -1.46, R) >consensus UCAUUUUCCUUUGAGGCAGUUGAUUUCGCCAACUGCAAGGAAAAUAUUUGAAAUUUACGAGUAUUGGCAAUUGUUUGAUAUUUUACCAGCGCUACGAAUUUGUAUUCG ..(((((((((....(((((((.......))))))))))))))))....(((...(((((((..((((......................))))...)))))))))). (-23.61 = -23.37 + -0.24)

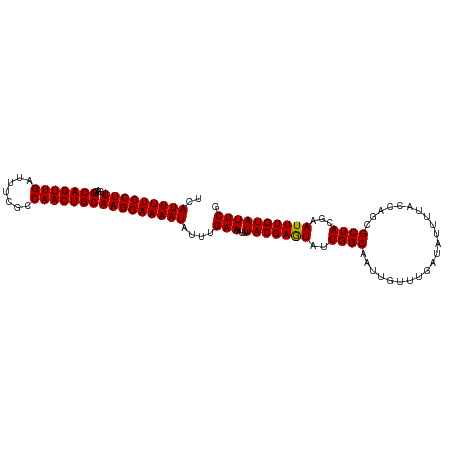
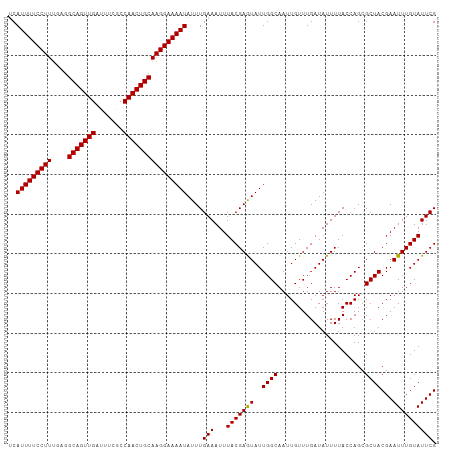
| Location | 6,586,019 – 6,586,127 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 97.59 |
| Shannon entropy | 0.04034 |
| G+C content | 0.34444 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -24.72 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6586019 108 - 22422827 CGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGCGAAAUCAACUGCCUCAAAGGAAAAUGC (((((....)))))..(((.((((((............)))))).....)))..........((((((((((((((((((.....))))))))....)))))))))). ( -24.90, z-score = -2.96, R) >droSec1.super_4 5971733 108 + 6179234 CGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUAUUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGCGAAAUCAACUGCCUCAAAGGAAAAUGA (((((....)))))..(((.((((((............)))))).....)))..........((((((((((((((((((.....))))))))....)))))))))). ( -25.20, z-score = -2.91, R) >droYak2.chrX 2549172 108 - 21770863 CGAAUACAAAUUCGUAGCACUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGAGAAAUCAACUGCCUCCAAGGAAAAUGA (((((....)))))......((((((............))))))..................(((((((((((((((((.(....))))))))....)))))))))). ( -25.00, z-score = -3.32, R) >droEre2.scaffold_4690 15512937 108 - 18748788 CGAAUACAAAUUCGUAGCACUGGUAAAAUAUCAAACAAUUGCCAAUAUUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGAGAAAUCAACUGCCUCAAAGGAAAAUGA (((((....)))))......((((((............))))))..................(((((((((((((((((.(....))))))))....)))))))))). ( -25.00, z-score = -3.35, R) >droSim1.chrX_random 2097937 108 - 5698898 CGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGCGAAAUCAACUGCCUCAAAGGAAAAUGA (((((....)))))..(((.((((((............)))))).....)))..........((((((((((((((((((.....))))))))....)))))))))). ( -25.20, z-score = -3.01, R) >consensus CGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUGGCGAAAUCAACUGCCUCAAAGGAAAAUGA (((((....)))))......((((((............))))))..................((((((((((((((((((.....))))))))....)))))))))). (-24.72 = -24.72 + 0.00)

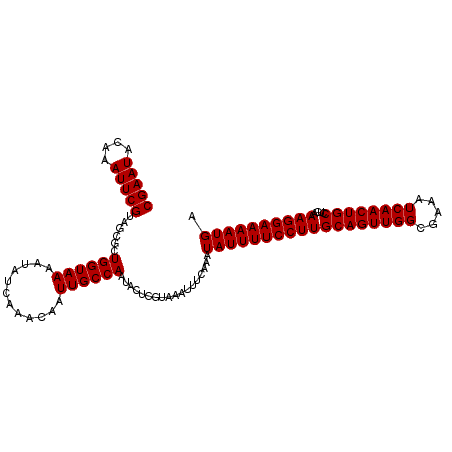
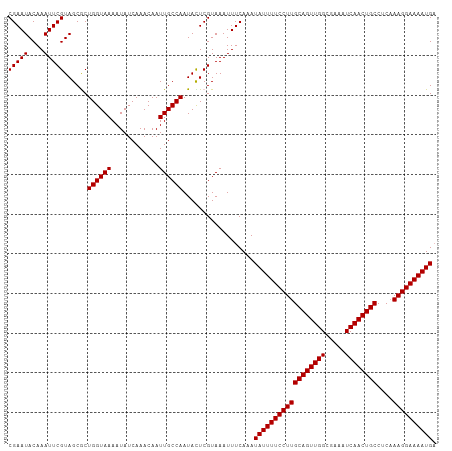
| Location | 6,586,048 – 6,586,155 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 98.13 |
| Shannon entropy | 0.03164 |
| G+C content | 0.32523 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6586048 107 - 22422827 UUUUUACAGUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((..((((.((((((((((((((((((....)))))..(((.((((((............)))))).....)))..)))))))))....))))))))..)) ( -22.20, z-score = -1.86, R) >droSec1.super_4 5971762 107 + 6179234 UUUUUGCAGUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUAUUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((..((((.(((((((((((((((((((.......(.....)((((((............)))))).))))))...)))))))))....))))))))..)) ( -22.10, z-score = -1.33, R) >droYak2.chrX 2549201 107 - 21770863 UUUUUGCAGUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCACUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((..((((.((((((((((((((((((....)))))......((((((............))))))..........)))))))))....))))))))..)) ( -21.60, z-score = -1.50, R) >droEre2.scaffold_4690 15512966 107 - 18748788 UUUUUGCAAUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCACUGGUAAAAUAUCAAACAAUUGCCAAUAUUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((((((((.((((((((((((((((((((......)))....((((((............))))))...))))...)))))))))....)))))))))))) ( -24.70, z-score = -2.77, R) >droSim1.chrX_random 2097966 107 - 5698898 UUUUUGCAGUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((..((((.((((((((((((((((((....)))))..(((.((((((............)))))).....)))..)))))))))....))))))))..)) ( -21.90, z-score = -1.23, R) >consensus UUUUUGCAGUUGCAGAGGAAUUUGGAAUCGAAUACAAAUUCGUAGCGCUGGUAAAAUAUCAAACAAUUGCCAAUACUCGUAAAUUUCAAAUAUUUUCCUUGCAGUUG ......((((((((.((((((((((((((((((....)))))......((((((............))))))..........)))))))))....)))))))))))) (-22.22 = -22.06 + -0.16)

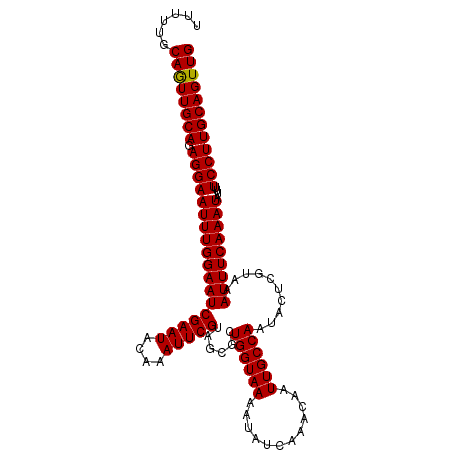
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:30 2011