
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,548,653 – 6,548,755 |
| Length | 102 |
| Max. P | 0.982343 |

| Location | 6,548,653 – 6,548,755 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.26 |
| Shannon entropy | 0.36997 |
| G+C content | 0.49009 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -18.76 |
| Energy contribution | -17.71 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
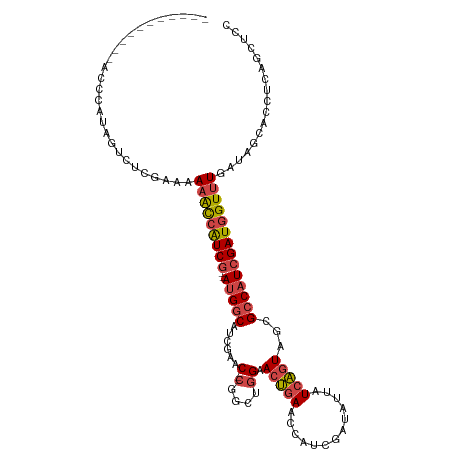
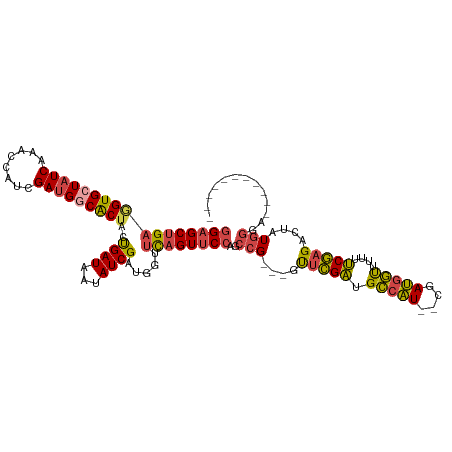
>dm3.chrX 6548653 102 + 22422827 -----------AUCCAUAAUCUCGAAAAAACCAUCG--AUGGCAUCGAAC---CGGCUGGAACUGAACCAUCGAUAUUAUCAGUAGCGCCAUCGAUGGUUUGAUAGCACCUCAGCUCC -----------................(((((((((--(((((..((...---))(((...(((((.............))))))))))))))))))))))...(((......))).. ( -30.32, z-score = -2.89, R) >droEre2.scaffold_4690 15475925 105 + 18748788 ---------CGACCCAUAGUCUCGAAAAAACCAUCGG-AUGGCAUCGAAC---CGGCUGGAACUGAACCGUCGAUAGUAUCAGUAGCGCCAUCGAUGGUUUGAUAGCACCUCAGCUCC ---------.(((.....)))......(((((((((.-(((((..((...---))(((...(((((..(.......)..))))))))))))))))))))))...(((......))).. ( -29.30, z-score = -0.92, R) >droYak2.chrX 2511258 103 + 21770863 ----------ACCCCAUAGUCUCGAAAAAACCAUCG--AUGGCAUCGAAC---CGGCUGGAACUGAACCAUCGAUAUUAUCAGUAGCGCCAUCGAUGGUUUGAUACCACCUCAGCUCC ----------.................(((((((((--(((((..((...---))(((...(((((.............))))))))))))))))))))))................. ( -28.92, z-score = -2.60, R) >droSec1.super_4 5922938 102 - 6179234 -----------UCCCAUUGUCUCGAAAAAACCAUCG--AUGGCAUCGAAC---CGGCUGGAACUGAACCAUCGAUAUUAUCAGUAGUGCCAUCGAUGGUUUGAUAGCACAUCAGCUCC -----------................(((((((((--(((((((....(---(....)).(((((.............))))).))))))))))))))))...(((......))).. ( -31.52, z-score = -2.81, R) >droSim1.chrX 5151674 102 + 17042790 -----------UCCCAUAGUCUCGAAAAAACCAUCG--AUGGCAUCGAAC---CGGCUGGAACUGAACCAUCGAUAUUAUCAGUAGUGCCAUCGAUGGUUUGAUAGCACCUCAGCUCC -----------................(((((((((--(((((((....(---(....)).(((((.............))))).))))))))))))))))...(((......))).. ( -31.52, z-score = -2.99, R) >droPer1.super_0 3818545 118 - 11822988 AACCUUGGGCUCGAUAAAAUCUUGGCUAUGUGGUCAGGACGACACCAGGCGUUCCAUCCGAACCAAAAUAUCGAUACUAUCGAUAGUGCCCUCGAUGGUUUGAUACCACUACCGCGCC .....(((..(((......((((((((....)))))))))))..)))(((((((.....)))).....((((((.((((((((........))))))))))))))..........))) ( -32.60, z-score = -1.09, R) >consensus ___________ACCCAUAGUCUCGAAAAAACCAUCG__AUGGCAUCGAAC___CGGCUGGAACUGAACCAUCGAUAUUAUCAGUAGCGCCAUCGAUGGUUUGAUAGCACCUCAGCUCC .....................((((.....((((....))))..))))......((((((...((((((((((((...............))))))))))))........)))))).. (-18.76 = -17.71 + -1.05)
| Location | 6,548,653 – 6,548,755 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Shannon entropy | 0.36997 |
| G+C content | 0.49009 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -29.67 |
| Energy contribution | -29.65 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6548653 102 - 22422827 GGAGCUGAGGUGCUAUCAAACCAUCGAUGGCGCUACUGAUAAUAUCGAUGGUUCAGUUCCAGCCG---GUUCGAUGCCAU--CGAUGGUUUUUUCGAGAUUAUGGAU----------- (((((((((((...)))..((((((((((...(....)....))))))))))))))))))..(((---.(((((.((((.--...))))....)))))....)))..----------- ( -35.30, z-score = -2.21, R) >droEre2.scaffold_4690 15475925 105 - 18748788 GGAGCUGAGGUGCUAUCAAACCAUCGAUGGCGCUACUGAUACUAUCGACGGUUCAGUUCCAGCCG---GUUCGAUGCCAU-CCGAUGGUUUUUUCGAGACUAUGGGUCG--------- (((((((((((((((((........)))))))))((((..........))))))))))))....(---((.....)))..-((.((((((((...)))))))).))...--------- ( -36.90, z-score = -1.79, R) >droYak2.chrX 2511258 103 - 21770863 GGAGCUGAGGUGGUAUCAAACCAUCGAUGGCGCUACUGAUAAUAUCGAUGGUUCAGUUCCAGCCG---GUUCGAUGCCAU--CGAUGGUUUUUUCGAGACUAUGGGGU---------- .((((((.(.(((.((..(((((((((((...(....)....)))))))))))..)).))).)))---))))...(((..--(.((((((((...)))))))).))))---------- ( -36.20, z-score = -1.91, R) >droSec1.super_4 5922938 102 + 6179234 GGAGCUGAUGUGCUAUCAAACCAUCGAUGGCACUACUGAUAAUAUCGAUGGUUCAGUUCCAGCCG---GUUCGAUGCCAU--CGAUGGUUUUUUCGAGACAAUGGGA----------- ((((((((((......)).((((((((((.((....))....))))))))))))))))))..(((---.(((((.((((.--...))))....)))))....)))..----------- ( -35.20, z-score = -2.33, R) >droSim1.chrX 5151674 102 - 17042790 GGAGCUGAGGUGCUAUCAAACCAUCGAUGGCACUACUGAUAAUAUCGAUGGUUCAGUUCCAGCCG---GUUCGAUGCCAU--CGAUGGUUUUUUCGAGACUAUGGGA----------- (((((((((((((((((........)))))))))..((((...)))).....))))))))..(((---.(((((.((((.--...))))....)))))....)))..----------- ( -35.70, z-score = -2.32, R) >droPer1.super_0 3818545 118 + 11822988 GGCGCGGUAGUGGUAUCAAACCAUCGAGGGCACUAUCGAUAGUAUCGAUAUUUUGGUUCGGAUGGAACGCCUGGUGUCGUCCUGACCACAUAGCCAAGAUUUUAUCGAGCCCAAGGUU (((.((((((((((.....))))((...(((..(((((((...)))))))...(((((.(((((..((.....))..))))).)))))....)))..))..)))))).)))....... ( -39.30, z-score = -1.68, R) >consensus GGAGCUGAGGUGCUAUCAAACCAUCGAUGGCACUACUGAUAAUAUCGAUGGUUCAGUUCCAGCCG___GUUCGAUGCCAU__CGAUGGUUUUUUCGAGACUAUGGGA___________ (((((((((((((((((........)))))))))..((((...)))).....))))))))..(((....(((((.(((((....)))))....)))))....)))............. (-29.67 = -29.65 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:28 2011