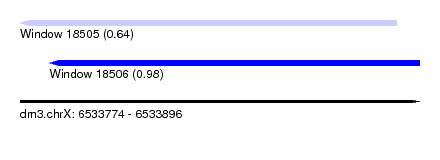
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,533,774 – 6,533,896 |
| Length | 122 |
| Max. P | 0.984641 |

| Location | 6,533,774 – 6,533,889 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Shannon entropy | 0.15632 |
| G+C content | 0.38921 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.40 |
| Energy contribution | -24.36 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6533774 115 - 22422827 ----UUUUUGGUCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCUGAGAGAUUUCCUUAAAUGGACCCCAAAACGAUCGGGUUGUUAAUGGCCAAGUUAAAUUAGGG ----..((((((((....(((.((((....((..(((((............)))))..)).(((((((......))).........)))))))).)))..))))))))........... ( -25.00, z-score = -0.17, R) >droEre2.scaffold_4690 15461046 114 - 18748788 ----UUUUUGGUCAGCCAAAUUAUGCCAAGUUUGGCUAAAUAGCUUCAAAAUUAGCAGAGUGAUUUCCUUAAAUGGACUCC-AAACGAUCGGGUUGCUAAUGACCAAGUUAAAUUAGGG ----..((((((((((((((((......))))))))).............((((((((..((((((((......)))....-....)))))..)))))))))))))))........... ( -33.20, z-score = -2.98, R) >droYak2.chrX 2496830 115 - 21770863 ---UUUUUUGGUCAGCCAAAUUACCCCAAAAUUGGCUAAAUAGCUUCAAAAUUAGCAGAGUGAUUUCCUUUAAUGGACGCC-AAACGGUCGGGUUGUUAAUGACCAAGUUAAAUUAGGG ---...((((((((....(((.((((.....(((((......(((........))).........(((......))).)))-))......)))).)))..))))))))........... ( -27.90, z-score = -1.11, R) >droSec1.super_4 5908347 119 + 6179234 AUAUUUCUUGGCCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCAGAGAGAUUUCCUUAAAUGGACCCCGAAACGAUCGGGUUGUUAAUGGCCAAGUUAAAUUAGGG ......(((((((((((((.(((....((((((.(((((............)))))....))))))...))).)))))(((((.....))))).......))))))))........... ( -31.50, z-score = -1.74, R) >droSim1.chrX 5136108 115 - 17042790 ----UUCUUGGCCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCAGAGAGAUUUCCUUAAAUGGACCCCGAAACGAUCGGGUUGUUAAUGGCCAAGUUAAAUUAGGG ----..(((((((((((((.(((....((((((.(((((............)))))....))))))...))).)))))(((((.....))))).......))))))))........... ( -31.50, z-score = -1.84, R) >consensus ____UUUUUGGUCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCAGAGAGAUUUCCUUAAAUGGACCCC_AAACGAUCGGGUUGUUAAUGGCCAAGUUAAAUUAGGG ......(((((((((((.((((......)))).)))).............((((((((...(((((((......))).........))))...)))))))))))))))........... (-25.40 = -24.36 + -1.04)

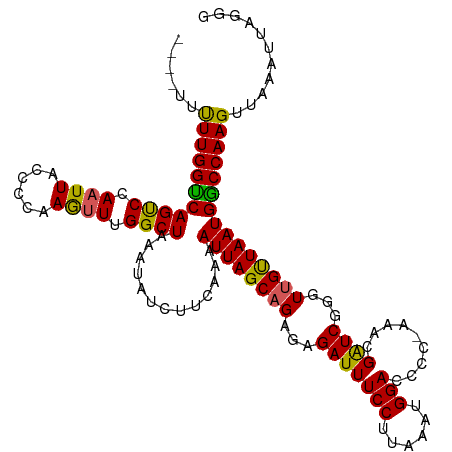
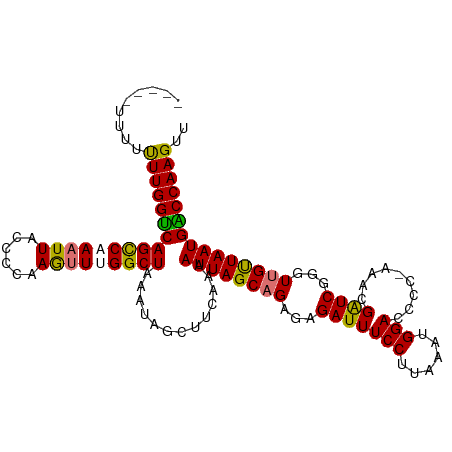
| Location | 6,533,783 – 6,533,896 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.35 |
| Shannon entropy | 0.20011 |
| G+C content | 0.37819 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6533783 113 - 22422827 UAUAUUUUUUUUGGUCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCUGAGAGAUUUCCUUAAAUGGACCCCAAAACGAUCGGGUUGUUAAUGGCCAAGUU .........((((((((....(((.((((....((..(((((............)))))..)).(((((((......))).........)))))))).)))..)))))))).. ( -25.00, z-score = -0.96, R) >droEre2.scaffold_4690 15461055 106 - 18748788 ------UUUUUUGGUCAGCCAAAUUAUGCCAAGUUUGGCUAAAUAGCUUCAAAAUUAGCAGAGUGAUUUCCUUAAAUGGACUCC-AAACGAUCGGGUUGCUAAUGACCAAGUU ------...((((((((((((((((......))))))))).............((((((((..((((((((......)))....-....)))))..))))))))))))))).. ( -33.20, z-score = -3.68, R) >droYak2.chrX 2496839 107 - 21770863 -----UUUUUUUGGUCAGCCAAAUUACCCCAAAAUUGGCUAAAUAGCUUCAAAAUUAGCAGAGUGAUUUCCUUUAAUGGACGCC-AAACGGUCGGGUUGUUAAUGACCAAGUU -----....((((((((....(((.((((.....(((((......(((........))).........(((......))).)))-))......)))).)))..)))))))).. ( -27.90, z-score = -1.87, R) >droSim1.chrX 5136117 113 - 17042790 UAUAUAUUUCUUGGCCAGUCCAAUUACCCCAAGUUUGGCUAAAUAUCUUCAAAAUUAGCAGAGAGAUUUCCUUAAAUGGACCCCGAAACGAUCGGGUUGUUAAUGGCCAAGUU .........(((((((((((((.(((....((((((.(((((............)))))....))))))...))).)))))(((((.....))))).......)))))))).. ( -31.50, z-score = -2.80, R) >consensus _____UUUUUUUGGUCAGCCAAAUUACCCCAAGUUUGGCUAAAUAGCUUCAAAAUUAGCAGAGAGAUUUCCUUAAAUGGACCCC_AAACGAUCGGGUUGUUAAUGACCAAGUU .........((((((((((((((((......))))))))).............((((((((...(((((((......))).........))))...))))))))))))))).. (-26.79 = -26.35 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:25 2011