| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,513,524 – 6,513,630 |
| Length | 106 |
| Max. P | 0.649917 |

| Location | 6,513,524 – 6,513,630 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.70 |
| Shannon entropy | 0.52652 |
| G+C content | 0.41790 |
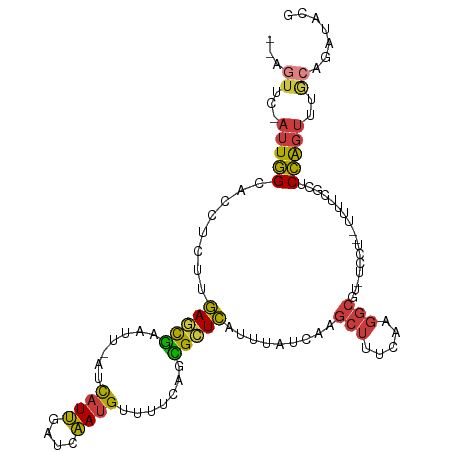
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -11.98 |
| Energy contribution | -11.68 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.649917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
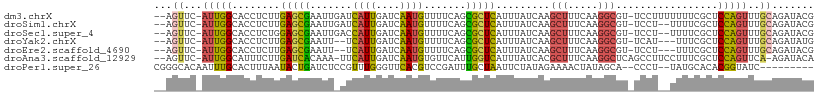
>dm3.chrX 6513524 106 - 22422827 --AGUUC-AUUGGCACCUCUUGAGCGAAUUGAUCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU-UCCUUUUUUUCGCUCCAGUUUGCAGAUACG --.....-(((.(((...((.(((((((((((((...)))))))......((((((..................)))))-)........)))))).))..))).)))... ( -23.37, z-score = -1.00, R) >droSim1.chrX 5118124 104 - 17042790 --AGUUC-AUUGGCACCUCUUGAGCGAAUUGAUCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU-UCCU--UUUUCGCUCCAGUUUGCAGAUACG --.....-(((.(((...((.(((((((((((((...)))))))......((((((..................)))))-)...--...)))))).))..))).)))... ( -23.37, z-score = -0.90, R) >droSec1.super_4 5888188 104 + 6179234 --AGUUC-AUUGGCACCUCUGGAGCGAAUUGACCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU-UCCU--UUUUCGCUCCAGUUUGCAGAUACG --.....-(((.(((...((((((((((..((.(.(((((.((((...........)))).)))))(((.....)))).-))..--..))))))))))..))).)))... ( -28.60, z-score = -2.33, R) >droYak2.chrX 2476785 101 - 21770863 --AGUUC-AUUGGCACCUCUUGAGCGAAUU--UCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU-UCAU---UUUCGCUCCAGUUUGCAGAUAUG --.....-(((.(((...((.(((((((..--...(((((.((((...........)))).)))))(((.....)))..-....---.))))))).))..))).)))... ( -22.10, z-score = -1.02, R) >droEre2.scaffold_4690 15440859 101 - 18748788 --AGUUC-AUUGGCACCUCUUGAGCGAAUU--UCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU-UCCU---UUUCGCUCCAGUUUGCAGAUACG --.....-(((.(((...((.(((((((..--...(((((.((((...........)))).)))))(((.....)))..-....---.))))))).))..))).)))... ( -22.10, z-score = -1.15, R) >droAna3.scaffold_12929 3082330 105 - 3277472 --AGUUC-AUUGGCAUUUCUUGAUCACAAA-UUCAUUGAUCAAUGUGUUCAUUGGUCAUUUAUCACGCUUUCAAGGCUCAGCCUUCCUUUCGCUCCAGUUCA-AGAUACA --.....-.........((((((.(.....-.....(((((((((....)))))))))........((....(((((...)))))......))....).)))-))).... ( -21.00, z-score = -1.71, R) >droPer1.super_26 996917 97 + 1349181 CGGGCACAAUUUGCACUUUAAUACUGAUCUCCGUUUGGGUUCACGUCCGAUUUGCUAAUUCUAUAGAAAACUAUAGCA--CCCU--UAUGCACACGGUAUC--------- ...(((.....)))......((((((.....(((..((((((......))..........((((((....)))))).)--))).--.)))....)))))).--------- ( -16.20, z-score = 0.26, R) >consensus __AGUUC_AUUGGCACCUCUUGAGCGAAUU_AUCAUUGAUCAAUGUUUUCAGCGCUCAUUUAUCAAGCUUUCAAGGCGU_UCCU__UUUUCGCUCCAGUUUGCAGAUACG ...((...(((((........(((((.......((((....)))).......))))).........(((.....))).................)))))..))....... (-11.98 = -11.68 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:19 2011