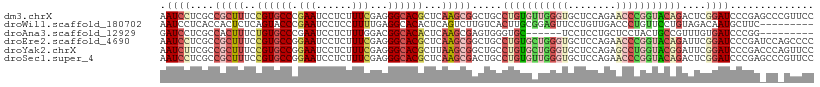
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,472,491 – 6,472,600 |
| Length | 109 |
| Max. P | 0.969857 |

| Location | 6,472,491 – 6,472,600 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.53 |
| Shannon entropy | 0.55704 |
| G+C content | 0.60006 |
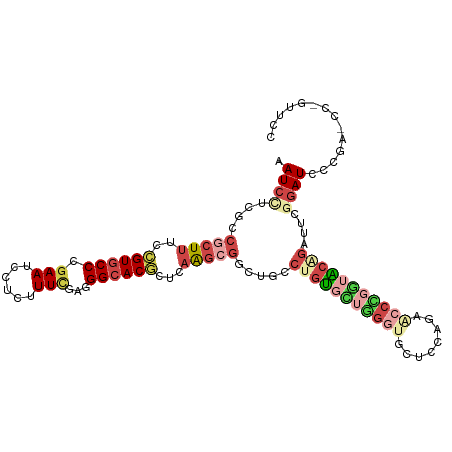
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -20.74 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
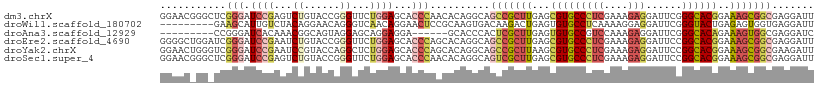
>dm3.chrX 6472491 109 + 22422827 GGAACGGGCUCGGGAUCCGAGUCUGUACCGGGUUCUGGAGCACCCAACACAGGCAGCCGCUUGAGCGUGCCCUCGAAAGAGGAUUCGGGCACGGAAAGCGGCGAGGAUU ((.((((((((((...)))))))))).))((((.(....).))))..........(((((((...(((((((((....))......)))))))..)))))))....... ( -53.20, z-score = -3.47, R) >droWil1.scaffold_180702 4396632 100 + 4511350 ---------GAAGCAUUGUCUACAGGAACAGGGUCAACAGGAACUCCGCAAGUGACAAGACUGAGUGUGCCUCAAAAGGAGGAUUCGGGUACUGAGAGUGGUGAGGAUU ---------....((((((((.(((..((...((((...((....)).....))))....((((((...((((.....)))))))))))).)))))).)))))...... ( -23.40, z-score = 0.76, R) >droAna3.scaffold_12929 3031827 94 + 3277472 ---------CCGGGAUCACAAACGGCAGUAGGAGCAGGAGGA------GCACCCACUCGCUUGAGUGUGCCGUCCAAAGAGGAUUCGGGCACAGAAAGUGGCGAGGAUC ---------..(((.((.....(.((.......)).)...))------...))).((((((....((((((((((.....))))...))))))......)))))).... ( -28.60, z-score = -0.44, R) >droEre2.scaffold_4690 15403749 109 + 18748788 GGGGCUGGAUCGGGAUCCGAAUCUGUACCGGGUUCUGGAGCACCCAGCACAGGCAGCCGCUUGAGCGUGCCCUCGAAAGAGGAUUCCGGCACGGAAAGCGGCGAGGAUU ((((((...((((((((((.........))))))))))))).))).((....)).(((((((...(((((((((....)))(....)))))))..)))))))....... ( -50.40, z-score = -2.32, R) >droYak2.chrX 2439137 109 + 21770863 GGAACUGGGUCGGGAUCCGAAUCCGUACCAGGCUCUGGAGCACCCAGCACAGGCAGCCGCUUAAGCGUGCCCUCGAAAGAGGAUUCCGGCACGGAAAGCGGCGAAGAUU ....((((((..((((....))))((.((((...)))).))))))))........(((((((...(((((((((....)))(....)))))))..)))))))....... ( -45.10, z-score = -1.90, R) >droSec1.super_4 5850621 109 - 6179234 GGAACGGGCUCGGGAUCCGAGUCUGUACCGGGUUCUGGAGCACCCAACACAGGCAGUCGCUUGAGCGUGCCCUCGAAAGAGGAUUCCGGCACGGAAAGCGGCGAGGAUU ((.((((((((((...)))))))))).))((((.(....).))))..........(((((((...(((((((((....)))(....)))))))..)))))))....... ( -49.90, z-score = -2.67, R) >consensus GGAAC_GG_UCGGGAUCCGAAUCUGUACCAGGUUCUGGAGCACCCAACACAGGCAGCCGCUUGAGCGUGCCCUCGAAAGAGGAUUCCGGCACGGAAAGCGGCGAGGAUU ...........(((.((((...((......))...))))...)))..........(((((((...(((((((((....)))......))))))..)))))))....... (-20.74 = -22.55 + 1.81)
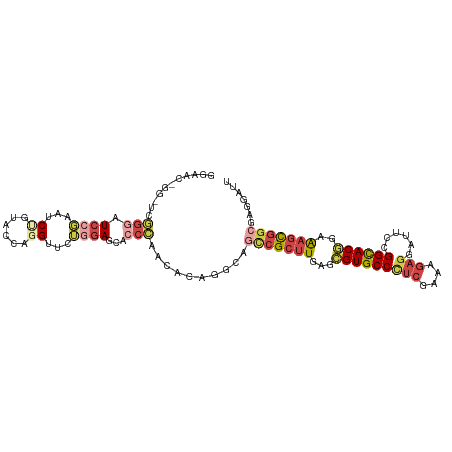
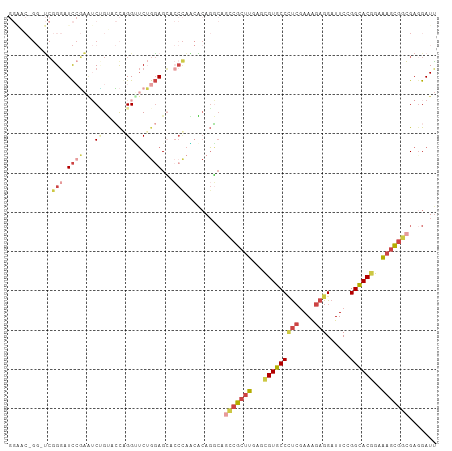
| Location | 6,472,491 – 6,472,600 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.53 |
| Shannon entropy | 0.55704 |
| G+C content | 0.60006 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -19.93 |
| Energy contribution | -22.43 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6472491 109 - 22422827 AAUCCUCGCCGCUUUCCGUGCCCGAAUCCUCUUUCGAGGGCACGCUCAAGCGGCUGCCUGUGUUGGGUGCUCCAGAACCCGGUACAGACUCGGAUCCCGAGCCCGUUCC .......(((((((..((((((((((......)))..)))))))...)))))))...(((((..((((........))))..))))).(((((...)))))........ ( -47.30, z-score = -3.49, R) >droWil1.scaffold_180702 4396632 100 - 4511350 AAUCCUCACCACUCUCAGUACCCGAAUCCUCCUUUUGAGGCACACUCAGUCUUGUCACUUGCGGAGUUCCUGUUGACCCUGUUCCUGUAGACAAUGCUUC--------- .................((.((((((.......)))).)).))....(((.(((((((..((((.(((......))).))))....)).))))).)))..--------- ( -17.00, z-score = 0.78, R) >droAna3.scaffold_12929 3031827 94 - 3277472 GAUCCUCGCCACUUUCUGUGCCCGAAUCCUCUUUGGACGGCACACUCAAGCGAGUGGGUGC------UCCUCCUGCUCCUACUGCCGUUUGUGAUCCCGG--------- ((((............((((((....(((.....))).))))))..((((((((((((.((------.......)).))))))..)))))).))))....--------- ( -30.60, z-score = -2.42, R) >droEre2.scaffold_4690 15403749 109 - 18748788 AAUCCUCGCCGCUUUCCGUGCCGGAAUCCUCUUUCGAGGGCACGCUCAAGCGGCUGCCUGUGCUGGGUGCUCCAGAACCCGGUACAGAUUCGGAUCCCGAUCCAGCCCC .......(((((((..((((((.(((......)))...))))))...))))))).(((((((((((((........)))))))))))....((((....)))).))... ( -48.10, z-score = -3.33, R) >droYak2.chrX 2439137 109 - 21770863 AAUCUUCGCCGCUUUCCGUGCCGGAAUCCUCUUUCGAGGGCACGCUUAAGCGGCUGCCUGUGCUGGGUGCUCCAGAGCCUGGUACGGAUUCGGAUCCCGACCCAGUUCC .......(((((((..((((((.(((......)))...))))))...)))))))...((((((..(((........)))..))))))..((((...))))......... ( -42.80, z-score = -1.39, R) >droSec1.super_4 5850621 109 + 6179234 AAUCCUCGCCGCUUUCCGUGCCGGAAUCCUCUUUCGAGGGCACGCUCAAGCGACUGCCUGUGUUGGGUGCUCCAGAACCCGGUACAGACUCGGAUCCCGAGCCCGUUCC .........(((((..((((((.(((......)))...))))))...))))).....(((((..((((........))))..))))).(((((...)))))........ ( -37.30, z-score = -0.54, R) >consensus AAUCCUCGCCGCUUUCCGUGCCCGAAUCCUCUUUCGAGGGCACGCUCAAGCGGCUGCCUGUGCUGGGUGCUCCAGAACCCGGUACAGAUUCGGAUCCCGA_CC_GUUCC .((((....(((((..((((((.(((......)))...))))))...))))).....(((((((((((........)))))))))))....)))).............. (-19.93 = -22.43 + 2.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:08 2011