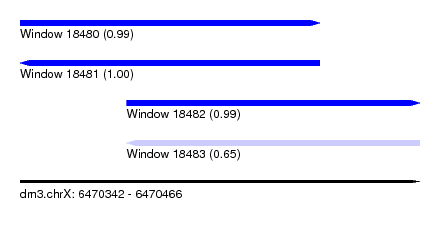
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,470,342 – 6,470,466 |
| Length | 124 |
| Max. P | 0.998081 |

| Location | 6,470,342 – 6,470,435 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.62473 |
| G+C content | 0.43387 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
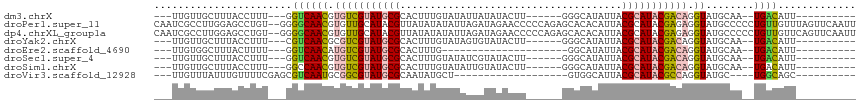
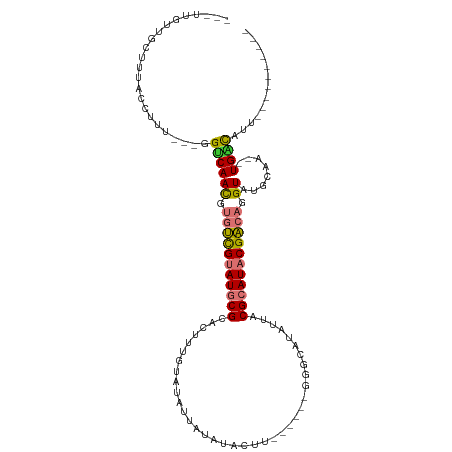
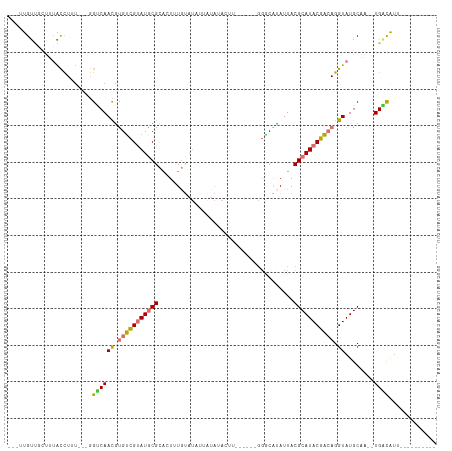
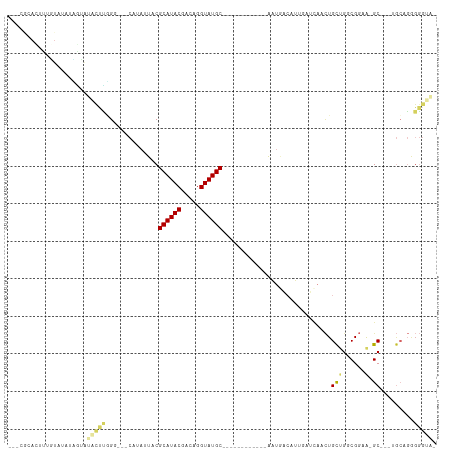
>dm3.chrX 6470342 93 + 22422827 ---UUGUUGCUUUACCUUU---GGUCAACGUGUCGUAUGCGCACUUUGUAUAUUAUAUACUU------GGGCAUAUUACGCAUACGACAGGUAUGCAA--UGACAUU---------- ---(..((((..((((...---(.....).((((((((((((.((..(((((...)))))..------)))).......)))))))))))))).))))--..)....---------- ( -28.51, z-score = -2.89, R) >droPer1.super_11 2774624 115 - 2846995 CAAUCGCCUUGGAGCCUGU--GGGGCAACGUGUUGCAUACGUUAUAUAUAUUAGAUAGAACCCCCAGAGCACACAUUACGCAUACGAGAGGUAUGCCCCCUGUUGUUUAGUUCAAUU ...........(((((((.--((((.(((((((...)))))))...(((.....)))...)))))))((((.(((....((((((.....))))))....)))))))..)))).... ( -27.00, z-score = -0.06, R) >dp4.chrXL_group1a 5618510 115 - 9151740 CAAUCGCCUUGGAGCCUGU--GGGGCAACGUGUUGCAUACGUUAUAUAUAUUAGAUAGAACCCCCAGAGCACACAUUACGCAUACGAGAGGUAUGCCCCCUGUUGUUCAGUUCAAUU ...........((((...(--((((.(((((((...)))))))...(((.....)))....)))))(((((.(((....((((((.....))))))....)))))))).)))).... ( -28.60, z-score = -0.44, R) >droYak2.chrX 2436952 93 + 21770863 ---UUGUUGCUUUACCUUU---CGUCAACGCGUCGUAUGCGCACUUUGUAUAGUGUAUACUU------GGGCAUAUUACGCAUACGACAGGUAUGCAA--UGACAUU---------- ---(..((((..(((((.(---(((....((((.((((((.((....(((((...))))).)------).)))))).))))..)))).))))).))))--..)....---------- ( -33.20, z-score = -3.75, R) >droEre2.scaffold_4690 15401580 78 + 18748788 ---UUGUGGCUUUACUUUU---GGUCAACAUGUCGUAUGCGCACUUUG---------------------GGCAUAUUACGCAUACGACAGGUAUGCAA--UGACAUU---------- ---...(((((........---)))))..(((((((((((.(.....)---------------------.))))))...((((((.....))))))..--.))))).---------- ( -23.90, z-score = -2.65, R) >droSec1.super_4 5848514 93 - 6179234 ---UUGUUGCUUUACCUUU---GGUCAACGUGUCGUAUGCGCACUUUGUAUAUCGUAUACUU------GGGCAUAUUACGCAUACGACAGGUAUGCAA--UGACAUU---------- ---(..((((..((((...---(.....).((((((((((((.((..(((((...)))))..------)))).......)))))))))))))).))))--..)....---------- ( -28.51, z-score = -2.40, R) >droSim1.chrX 5077217 93 + 17042790 ---UUGUUGCUUUACCUUU---GGCCAACGUGUCGUAUGCGCACUUUGUAUAUUGUAUACUU------GGGCAUAUUACGCAUACGACAGGUAUGCAA--UGACAUU---------- ---(..((((..((((...---(.....).((((((((((((.((..(((((...)))))..------)))).......)))))))))))))).))))--..)....---------- ( -28.51, z-score = -2.22, R) >droVir3.scaffold_12928 505520 81 + 7717345 ---UUGUUUAUUUGUUUUCGAGCGUCAAUGCGGCGUAUGCGCAAUAUGCU-------------------GUGGCAUUACGCAUACGCCAGGUAUGC----UGGCAGC---------- ---..................((((((((((((((((((((....((((.-------------------...))))..))))))))))..))))..----)))).))---------- ( -27.70, z-score = -0.64, R) >consensus ___UUGUUGCUUUACCUUU___GGUCAACGUGUCGUAUGCGCACUUUGUAUAUUAUAUACUU______GGGCAUAUUACGCAUACGACAGGUAUGCAA__UGACAUU__________ .......................((((((.(((((((((((.....................................))))))))))).))........))))............. (-14.52 = -14.57 + 0.05)
| Location | 6,470,342 – 6,470,435 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.62473 |
| G+C content | 0.43387 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.62 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6470342 93 - 22422827 ----------AAUGUCA--UUGCAUACCUGUCGUAUGCGUAAUAUGCCC------AAGUAUAUAAUAUACAAAGUGCGCAUACGACACGUUGACC---AAAGGUAAAGCAACAA--- ----------.......--((((.(((((((((((((((((((((((..------..))))))...........))))))))))))..(....).---..)))))..))))...--- ( -26.80, z-score = -3.07, R) >droPer1.super_11 2774624 115 + 2846995 AAUUGAACUAAACAACAGGGGGCAUACCUCUCGUAUGCGUAAUGUGUGCUCUGGGGGUUCUAUCUAAUAUAUAUAACGUAUGCAACACGUUGCCCC--ACAGGCUCCAAGGCGAUUG (((((.............((((((.((.....((((((((.(((((((...((((.......)))).))))))).)))))))).....))))))))--.............))))). ( -29.17, z-score = -0.10, R) >dp4.chrXL_group1a 5618510 115 + 9151740 AAUUGAACUGAACAACAGGGGGCAUACCUCUCGUAUGCGUAAUGUGUGCUCUGGGGGUUCUAUCUAAUAUAUAUAACGUAUGCAACACGUUGCCCC--ACAGGCUCCAAGGCGAUUG ..(((..(((.....)))((((((.((.....((((((((.(((((((...((((.......)))).))))))).)))))))).....))))))))--.)))(((....)))..... ( -30.90, z-score = -0.34, R) >droYak2.chrX 2436952 93 - 21770863 ----------AAUGUCA--UUGCAUACCUGUCGUAUGCGUAAUAUGCCC------AAGUAUACACUAUACAAAGUGCGCAUACGACGCGUUGACG---AAAGGUAAAGCAACAA--- ----------.......--((((.(((((((((((((((...(((((..------..)))))((((......)))))))))))))).((....))---..)))))..))))...--- ( -29.70, z-score = -3.12, R) >droEre2.scaffold_4690 15401580 78 - 18748788 ----------AAUGUCA--UUGCAUACCUGUCGUAUGCGUAAUAUGCC---------------------CAAAGUGCGCAUACGACAUGUUGACC---AAAAGUAAAGCCACAA--- ----------...((((--..(((....(((((((((((((...(...---------------------...).)))))))))))))))))))).---................--- ( -22.90, z-score = -3.21, R) >droSec1.super_4 5848514 93 + 6179234 ----------AAUGUCA--UUGCAUACCUGUCGUAUGCGUAAUAUGCCC------AAGUAUACGAUAUACAAAGUGCGCAUACGACACGUUGACC---AAAGGUAAAGCAACAA--- ----------.......--((((.(((((((((((((((((.(((((..------..)))))............))))))))))))..(....).---..)))))..))))...--- ( -25.62, z-score = -2.33, R) >droSim1.chrX 5077217 93 - 17042790 ----------AAUGUCA--UUGCAUACCUGUCGUAUGCGUAAUAUGCCC------AAGUAUACAAUAUACAAAGUGCGCAUACGACACGUUGGCC---AAAGGUAAAGCAACAA--- ----------.......--((((.(((((((((((((((((.(((((..------..)))))............))))))))))))..(....).---..)))))..))))...--- ( -25.92, z-score = -2.00, R) >droVir3.scaffold_12928 505520 81 - 7717345 ----------GCUGCCA----GCAUACCUGGCGUAUGCGUAAUGCCAC-------------------AGCAUAUUGCGCAUACGCCGCAUUGACGCUCGAAAACAAAUAAACAA--- ----------((.....----))......((((((((((((((((...-------------------.))))..))))))))))))((......))..................--- ( -27.20, z-score = -2.68, R) >consensus __________AAUGUCA__UUGCAUACCUGUCGUAUGCGUAAUAUGCCC______AAGUAUACAAUAUACAAAGUGCGCAUACGACACGUUGACC___AAAGGUAAAGCAACAA___ ......................((.((.(((((((((((((.................................))))))))))))).))))......................... (-15.72 = -15.62 + -0.09)
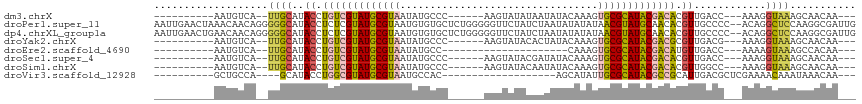
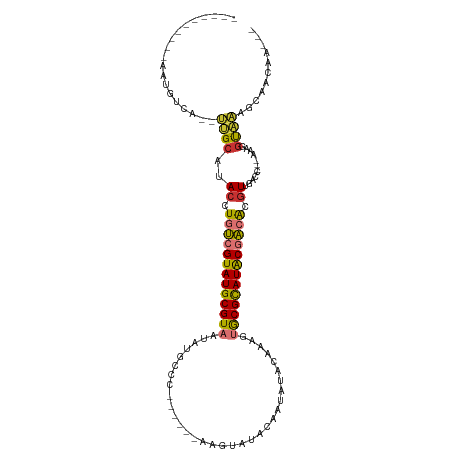
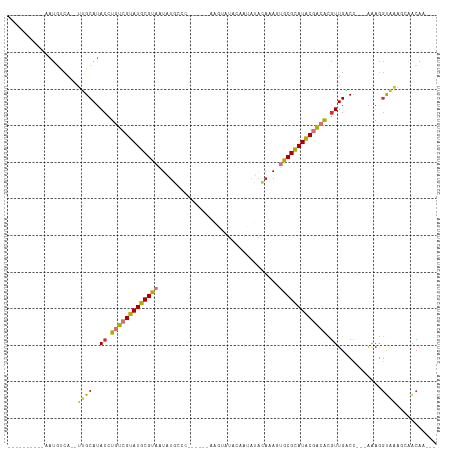
| Location | 6,470,375 – 6,470,466 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.50 |
| Shannon entropy | 0.76997 |
| G+C content | 0.47407 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
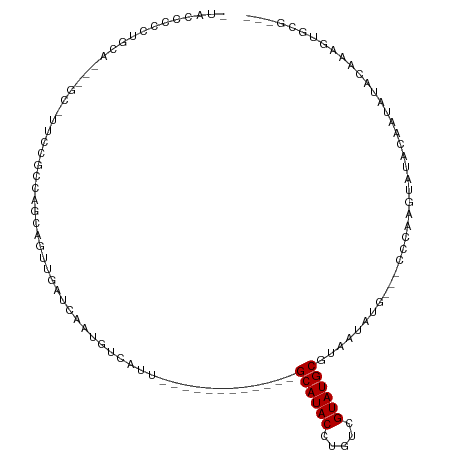
>dm3.chrX 6470375 91 + 22422827 ---CGCACUUUGUAUAUUAUAUACUUGGG---CAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUGGCGGAA-GC---UGCCGGGGGUA- ---.((.((..(((((...)))))..)))---).......((((((.....))))))------------..............(((.(((((((..-.)---)))))).))).- ( -28.30, z-score = -2.45, R) >droPer1.super_11 2774661 113 - 2846995 CGUUAUAUAUAUUAGAUAGAACCCCCAGAGCACACAUUACGCAUACGAGAGGUAUGCCCCCUGUUGUUUAGUUCAAUUAGUCAACUGCAGUCGGAA-GCAAUUGCAGAGGGGGA .....................(((((.(((((((((....((((((.....))))))....)).)))...))))..........((((((((....-)..))))))).))))). ( -33.30, z-score = -2.83, R) >dp4.chrXL_group1a 5618547 113 - 9151740 CGUUAUAUAUAUUAGAUAGAACCCCCAGAGCACACAUUACGCAUACGAGAGGUAUGCCCCCUGUUGUUCAGUUCAAUUAGUCAACUGCAGUCGGAA-GCAAUUGCAGAGGGGGA .....................(((((.(((((.(((....((((((.....))))))....))))))))...............((((((((....-)..))))))).))))). ( -35.70, z-score = -3.53, R) >droYak2.chrX 2436985 91 + 21770863 ---CGCACUUUGUAUAGUGUAUACUUGGG---CAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUGGCGGAA-GC---AGCAGGGGGUA- ---....(..(((....((((((((((.(---(.......))......)))))))))------------)...)))..)(((..((((((.(....-))---)))))..))).- ( -31.50, z-score = -3.39, R) >droEre2.scaffold_4690 15401613 76 + 18748788 -----------------CGCAC-UUUGGG---CAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUGGCGGAA-GC---AGCAGGGGGUA- -----------------.((.(-....))---).......((((((.....))))))------------..........(((..((((((.(....-))---)))))..))).- ( -25.30, z-score = -3.38, R) >droSec1.super_4 5848547 91 - 6179234 ---CGCACUUUGUAUAUCGUAUACUUGGG---CAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUGGCGGAA-GC---UGCAGGGGGUA- ---.((.((..(((((...)))))..)))---).......((((((.....))))))------------..........(((..((((.(((....-))---)))))..))).- ( -29.90, z-score = -2.89, R) >droSim1.chrX 5077250 91 + 17042790 ---CGCACUUUGUAUAUUGUAUACUUGGG---CAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUGAACUGCUGGCGGAA-GC---UGCAGGGGGUA- ---..(((..(((.(((((((((((((.(---(.......))......)))))))))------------)))))))..).))..((((.(((....-))---)))))......- ( -30.10, z-score = -3.22, R) >droMoj3.scaffold_6328 1465670 79 + 4453435 ---CCCAAUAUUUGCCUGUGGUUGUGUGC----GCAUUACGCAUACGCCAGGUAUGC-----------UGCAAGCAUACAUUG-UUGUUGGCGUUG-GC--------------- ---.(((((....(((..((((.((((((----(.....))))))))))).((((((-----------.....))))))....-.....)))))))-).--------------- ( -29.50, z-score = -1.11, R) >droVir3.scaffold_12928 505556 77 + 7717345 --------------CGCAAUAUGCUGUG-----GCAUUACGCAUACGCCAGGUAUGC-----------UGGCAGCAUUUAUU---UGUUGACGCUGUGCAAAUAAUUGGC---- --------------.((...((((....-----))))...))....(((((.....)-----------))))..((.(((((---(((.........)))))))).))..---- ( -21.40, z-score = 0.66, R) >consensus ___CGCACUUUGUAUAUAGUAUACUUGGG___CAUAUUACGCAUACGACAGGUAUGC____________AAUGACAUUGAUCAACUGCUGGCGGAA_GC___UGCAGGGGGUA_ .....................(((((..............((((((.....))))))...........................(((....)))..............))))). ( -8.10 = -8.94 + 0.84)

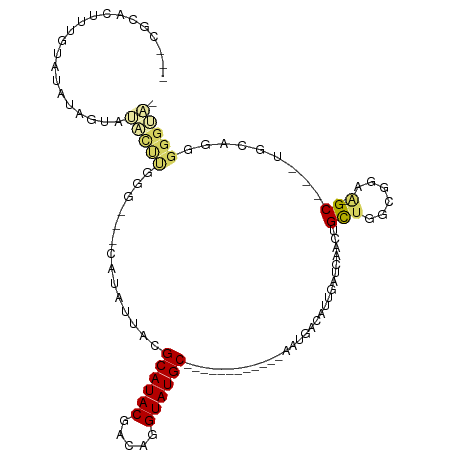
| Location | 6,470,375 – 6,470,466 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.50 |
| Shannon entropy | 0.76997 |
| G+C content | 0.47407 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -7.10 |
| Energy contribution | -7.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
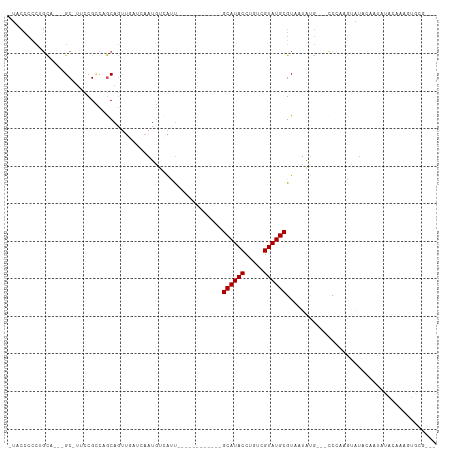
>dm3.chrX 6470375 91 - 22422827 -UACCCCCGGCA---GC-UUCCGCCAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUG---CCCAAGUAUAUAAUAUACAAAGUGCG--- -.......(((.---..-....))).(((.(((............------------((((((.....))))))...(((((---(....))))))......)))..))).--- ( -19.40, z-score = -0.86, R) >droPer1.super_11 2774661 113 + 2846995 UCCCCCUCUGCAAUUGC-UUCCGACUGCAGUUGACUAAUUGAACUAAACAACAGGGGGCAUACCUCUCGUAUGCGUAAUGUGUGCUCUGGGGGUUCUAUCUAAUAUAUAUAACG .(((((.((((((((((-........))))))).....(((.......))))))(((((((((....((....))....))))))))))))))..................... ( -32.90, z-score = -2.53, R) >dp4.chrXL_group1a 5618547 113 + 9151740 UCCCCCUCUGCAAUUGC-UUCCGACUGCAGUUGACUAAUUGAACUGAACAACAGGGGGCAUACCUCUCGUAUGCGUAAUGUGUGCUCUGGGGGUUCUAUCUAAUAUAUAUAACG .(((((....(((((((-........)))))))..........(((.....)))(((((((((....((....))....))))))))))))))..................... ( -33.60, z-score = -2.50, R) >droYak2.chrX 2436985 91 - 21770863 -UACCCCCUGCU---GC-UUCCGCCAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUG---CCCAAGUAUACACUAUACAAAGUGCG--- -......(((((---((-....).)))))).((((....))))..------------((((((.....))))))....((((---(....)))))((((......))))..--- ( -19.80, z-score = -1.27, R) >droEre2.scaffold_4690 15401613 76 - 18748788 -UACCCCCUGCU---GC-UUCCGCCAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUG---CCCAAA-GUGCG----------------- -......(((((---((-....).))))))(((....((((...(------------((((((.....)))))))..)))).---..))).-.....----------------- ( -16.60, z-score = -1.24, R) >droSec1.super_4 5848547 91 + 6179234 -UACCCCCUGCA---GC-UUCCGCCAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUG---CCCAAGUAUACGAUAUACAAAGUGCG--- -........(((---.(-((......(((((.(((....))))))------------)).....((((((((((........---.....))))))))))....)))))).--- ( -20.72, z-score = -1.55, R) >droSim1.chrX 5077250 91 - 17042790 -UACCCCCUGCA---GC-UUCCGCCAGCAGUUCAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUG---CCCAAGUAUACAAUAUACAAAGUGCG--- -......((((.---((-....))..)))).......((((...(------------((((((.....)))))))..))))(---((...(((((...)))))...).)).--- ( -15.70, z-score = -0.47, R) >droMoj3.scaffold_6328 1465670 79 - 4453435 ---------------GC-CAACGCCAACAA-CAAUGUAUGCUUGCA-----------GCAUACCUGGCGUAUGCGUAAUGC----GCACACAACCACAGGCAAAUAUUGGG--- ---------------((-(.((((((....-....((((((.....-----------)))))).)))))).((((.....)----)))..........)))..........--- ( -24.50, z-score = -1.12, R) >droVir3.scaffold_12928 505556 77 - 7717345 ----GCCAAUUAUUUGCACAGCGUCAACA---AAUAAAUGCUGCCA-----------GCAUACCUGGCGUAUGCGUAAUGC-----CACAGCAUAUUGCG-------------- ----((((......(((.((((((.....---.....))))))...-----------)))....))))....(((..((((-----....))))..))).-------------- ( -19.00, z-score = 0.10, R) >consensus _UACCCCCUGCA___GC_UUCCGCCAGCAGUUGAUCAAUGUCAUU____________GCAUACCUGUCGUAUGCGUAAUAUG___CCCAAGUAUACAAUAUACAAAGUGCG___ .........................................................((((((.....))))))........................................ ( -7.10 = -7.10 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:06 2011