| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,452,118 – 6,452,229 |
| Length | 111 |
| Max. P | 0.931479 |

| Location | 6,452,118 – 6,452,229 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.64505 |
| G+C content | 0.56128 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
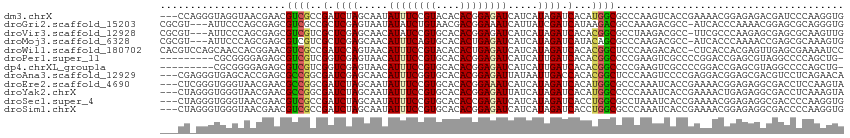
>dm3.chrX 6452118 111 + 22422827 ---CCAGGGUAGGUAACGAACGUCGCCGAUCUAGCAAUAUUUCCGUACACACGGAGAUCAUCAUAGAUCACAUGGCGCCCAAGUCACCGAAAACGGAGAGACGAUCCCAAGGUG ---((.((((.((..((....))..))((((((.....((((((((....)))))))).....)))))).......))))..(((.(((....)))...)))........)).. ( -34.90, z-score = -2.83, R) >droGri2.scaffold_15203 6915063 110 + 11997470 CGCGU---AUUCCCAGCGAGCGUCGCCGCUCGAGUAAUAUAUCUGUAACGACGGAAAUCAUUAUCGAUCAUAAGACGCCAAAGACGCC-AUCACCCAAAACGGAGCGCAGGGUG .....---...(((.(((.(((((...((((((.((((((.(((((....))))).)).))))))))((....)).))....))))).-.....((.....))..))).))).. ( -26.00, z-score = 0.40, R) >droVir3.scaffold_12928 491224 110 + 7717345 CGCGU---AUUCCCAGCGAGCGUCGUCGCUCGAGCAACAUAUCCGUGCACACGGAGAUCAUCAUAGAUCACACGGCGCCUAAGACGCC-UUCGCCCAAGAGCGAGCGCAAGUUG .....---.....((((..((((..((((((..((......(((((....)))))((((......))))....((((.......))))-...))....))))))))))..)))) ( -37.10, z-score = -1.90, R) >droMoj3.scaffold_6328 1450849 110 + 4453435 CGCGU---AUUCCCAGCGAGCGUCGUCGCUCGAGCAACAUUUCAGUGCACACUGAGAUCAUCAUAGAUCAUACAGCGCCCAAGACGCC-AUCACCCAAAACCGAGCGCAAAGUG .((((---........((((((....)))))).((......(((((....)))))((((......)))).....))............-...............))))...... ( -27.50, z-score = -1.80, R) >droWil1.scaffold_180702 4385384 113 + 4511350 CACGUCCAGCAACCACGGAACGUCGCCGAUCCAGUAACAUUUCCGUACACACUGAGAUCAUCAUAGAUCACACGGCUCCCAAGACACC-CUCACCACGAGUUGAGCGAAAAUCC ..((..((((......((...(((((((...((((.((......))....)))).((((......))))...))))......)))..)-)((.....))))))..))....... ( -21.30, z-score = -0.65, R) >droPer1.super_11 2763527 104 - 2846995 ---------CGCGGGGAGAGCGUCGUCGGUCGAGUAACAUUUCCGUGCACACGGAGAUCAUCAUUGAUCACACGGCCCCGAAGUCGCCCCGGACCGAGCGUAGGCCCCAGCUG- ---------.((((((...((((..((((((..((......(((((....)))))(((((....))))))).(((...((....))..)))))))))))))...)))).))..- ( -41.10, z-score = -1.44, R) >dp4.chrXL_group1a 5607105 104 - 9151740 ---------CGCGGGGAGAGCGUCGUCGGUCGAGUAACAUUUCCGUGCACACGGAGAUCAUCAUUGAUCACACGGCCCCGAAGUCGCCCCGGACCGAGCGUAGGCCCCAGCUG- ---------.((((((...((((..((((((..((......(((((....)))))(((((....))))))).(((...((....))..)))))))))))))...)))).))..- ( -41.10, z-score = -1.44, R) >droAna3.scaffold_12929 3018473 111 + 3277472 ---CGAGGGUGAGCACCGAGCGCCGGCGAUCGAGCAACAUUUCGGUGCACACGGAGAUUAUAAUUGACCACACGGCUCCCAAGUCCCCGAGGACGGAGCGACGUCCUCAGAACA ---...(((.((((((((((.....((......)).....))))))))....((((.(((....)))((....))))))....)))))(((((((......)))))))...... ( -40.30, z-score = -1.99, R) >droEre2.scaffold_4690 15392006 111 + 18748788 ---CUCGGGUGGGUAACGAACGCCGGCGAUCUAGCAAUAUUUCCGUGCACACGGAAAUCAUCAUAGAUCACAUGGCGCCCAAAUCACCGAAAACGGAGAGGCGACCUCCAAGUA ---.((((.((((.......((((((.((((((.....((((((((....)))))))).....)))))).).))))))))).....))))....((((.(....)))))..... ( -42.21, z-score = -3.86, R) >droYak2.chrX 2427813 111 + 21770863 ---CUAGGGUGGGUAACGAACGCCGGCGAUCUAGCAAUAUUUCCGUGCACACGGAGAUUAUCAUAGAUCACAUGGCCCCCAAAUCACCGAAAACUGAGAGGCGACCUCAAAGUA ---....(((((.........(((((.((((((.....((((((((....)))))))).....)))))).).)))).......)))))....(((..((((...))))..))). ( -35.99, z-score = -2.71, R) >droSec1.super_4 5838750 111 - 6179234 ---CUAGGGUGGGUAACGAACGUCGCCGAUCUAGCAAUAUUUCCGUGCACACCGAGAUCAUCAUAGAUCACCUGGCGCCUAAAUCACCGAAAACGGAGAGGCGACCCCAAGGUG ---(((((...((..((....))..))((((((.....(((((.((....)).))))).....)))))).)))))(((((......(((....)))..)))))(((....))). ( -33.80, z-score = -1.32, R) >droSim1.chrX 5066442 111 + 17042790 ---CUAGGGUGGGUAACGAACGUCGCCGAUCUAGCAAUAUUUCCGUGCACACGGAGAUCAUCAUAGAUCACCUGGCGCCCAAAUCACCGAAAACGGAGAGGCGACCCCAAGGUG ---(((((...((..((....))..))((((((.....((((((((....)))))))).....)))))).)))))((((....((.(((....))).))))))(((....))). ( -39.60, z-score = -2.71, R) >consensus ___CU_GGGUGCGCAACGAACGUCGCCGAUCGAGCAACAUUUCCGUGCACACGGAGAUCAUCAUAGAUCACACGGCGCCCAAGUCACCGAAAACCGAGAGGCGACCCCAAAGUG .....................((((..(.((((.....((((((((....)))))))).....)))).)...))))...................................... (-13.33 = -13.18 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:02 2011