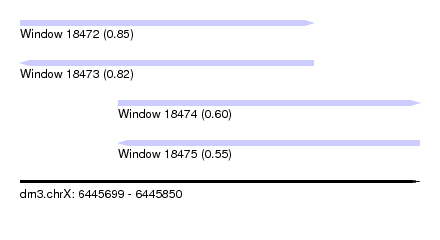
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,445,699 – 6,445,850 |
| Length | 151 |
| Max. P | 0.851058 |

| Location | 6,445,699 – 6,445,810 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.24116 |
| G+C content | 0.52506 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -26.45 |
| Energy contribution | -27.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6445699 111 + 22422827 ACUCCACUUGGACUCCACUCGCAGGCACUCAAACUUGAGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAC----ACA--UAACGAGUUGAGAGCGUAAAUGGG ...(((.(((..(((((((((.............((((((.((((....))))))))))..(((((......))))).......----...--...)))).)).)))..))).))). ( -33.60, z-score = -1.86, R) >droEre2.scaffold_4690 15385522 117 + 18748788 ACUCCACUUGGACUCCACUCGCAGGCACUCAAGCCUGAGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAUCUAUACAGAUACCGAGUUGAGAGCGAAAAUGGG ...(((..(((((((......(((((......))))).....)))))))((..((((....(((((......)))))....(((((....)))))..))))..))........))). ( -40.00, z-score = -2.68, R) >droYak2.chrX 2419797 96 + 21770863 ACUCCACUUGGACUCCACUCGCAGGCACUCAACGCUGAGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGCGCCUAUUUAC----CGAGAUACCGAG----------------- ......(((((.(((.......((((.(((.....(((((....)))))(((((((.....)).))))))).).))))......----.)))...)))))----------------- ( -24.54, z-score = -0.99, R) >droSec1.super_4 5832342 113 - 6179234 ACUCCACUUGGACUCCACUCGCAGGCACUCAAACUUGAGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAC----ACAGAUAUCGAGUUGAGAGCGUAAAUGGG ..(((....)))..(((.(..(..((.(((((.(((((((.((((....))))))......(((((......)))))(((((..----..))))))))))))))).)))..).))). ( -35.30, z-score = -1.98, R) >consensus ACUCCACUUGGACUCCACUCGCAGGCACUCAAACCUGAGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAC____ACAGAUACCGAGUUGAGAGCGUAAAUGGG ..(((....)))(((.(((((.((((.((((....))))..((((....))))))))....(((((......)))))...................))))).)))............ (-26.45 = -27.70 + 1.25)

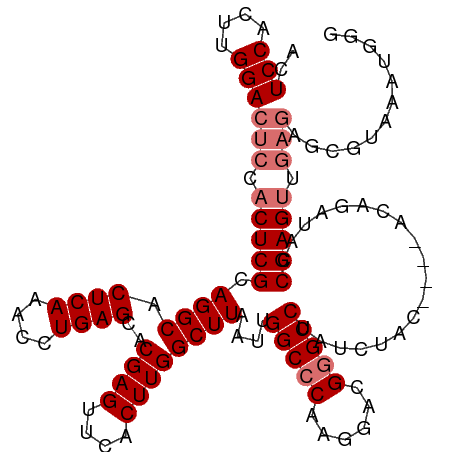
| Location | 6,445,699 – 6,445,810 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.24116 |
| G+C content | 0.52506 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -26.70 |
| Energy contribution | -29.95 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6445699 111 - 22422827 CCCAUUUACGCUCUCAACUCGUUA--UGU----GUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCUCAAGUUUGAGUGCCUGCGAGUGGAGUCCAAGUGGAGU .((((((..(((((..((((((..--((.----((.(((.((((((....)))))).)))..)))).........(..((((....))))..)..)))))))))))..))))))... ( -38.60, z-score = -2.88, R) >droEre2.scaffold_4690 15385522 117 - 18748788 CCCAUUUUCGCUCUCAACUCGGUAUCUGUAUAGAUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCUCAGGCUUGAGUGCCUGCGAGUGGAGUCCAAGUGGAGU .((((((..(((((..(((((.(((((((....)))))))((((((....)))))).....((((.((....)).).)))(((((......)))))))))))))))..))))))... ( -46.00, z-score = -3.55, R) >droYak2.chrX 2419797 96 - 21770863 -----------------CUCGGUAUCUCG----GUAAAUAGGCGCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCUCAGCGUUGAGUGCCUGCGAGUGGAGUCCAAGUGGAGU -----------------...((...((((----((.(((.(((.((....)).))).)))..)))......((((((((((......))))....)))))).))).))......... ( -26.60, z-score = 0.66, R) >droSec1.super_4 5832342 113 + 6179234 CCCAUUUACGCUCUCAACUCGAUAUCUGU----GUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCUCAAGUUUGAGUGCCUGCGAGUGGAGUCCAAGUGGAGU .((((((..(((((..(((((.(((((..----..)))))((((((....))))))...................(..((((....))))..)...))))))))))..))))))... ( -39.10, z-score = -2.74, R) >consensus CCCAUUUACGCUCUCAACUCGGUAUCUGU____GUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCUCAAGUUUGAGUGCCUGCGAGUGGAGUCCAAGUGGAGU .((((((..(((((..(((((...................((((((....))))))...................(..((((....))))..)...))))))))))..))))))... (-26.70 = -29.95 + 3.25)


| Location | 6,445,736 – 6,445,850 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.53 |
| Shannon entropy | 0.55436 |
| G+C content | 0.44668 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
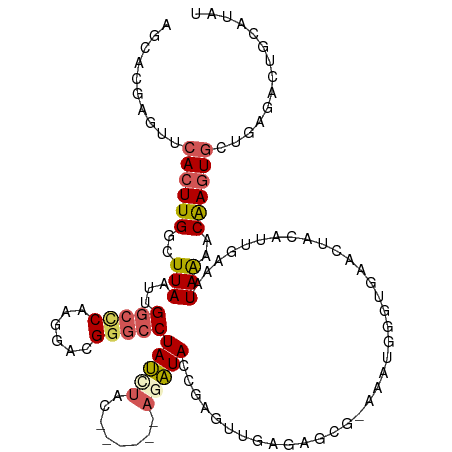
>dm3.chrX 6445736 114 + 22422827 AGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAC------ACAUAACGAGUUGAGAGCGUAAAUGGGUGAACUACAUUGGAAUAAAACAAGUGCUGAGACUGCAUAU (((((.((((((((((((((....(((((......)))))..((.((------.(.....).)).)))))).....))))))))))...(((........))))))))............ ( -30.90, z-score = -1.62, R) >droAna3.scaffold_12929 3012084 87 + 3277472 ----CGAGAUCACUUGGCUUAAUUGGCUCAAGGACGGAGCUAC----------ACGUACUUGGCA----------------GUAAUUUAUGGCAGUAGUUCGACUUCUGAUGCUUUC--- ----.((((((((((((((.....))).))))....(((((((----------.......((.((----------------........)).)))))))))......)))).)))..--- ( -19.61, z-score = 0.07, R) >droEre2.scaffold_4690 15385559 111 + 18748788 AGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAUCUAUACAGAUACCGAGUUGAGAGCGAAAAUGGGUGAUAUACAUUGAAGUAAAACAAGUGCAUAU--------- .((((..(((..((..((((....(((((......)))))....(((((....)))))..))))..)).((...((((.........))))...))..)))..))))....--------- ( -31.00, z-score = -2.33, R) >droYak2.chrX 2419834 99 + 21770863 AGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGCGCCUAUUUACC----GAGAUACCGAGU-----------------GAUCUACAUUGAAAUAACACAAGUGCUGAGACUGCAUAU (((((.((.((((((((.....((((....(((.....))).....))----))....))))))-----------------)).))...(((........))))))))............ ( -23.90, z-score = -1.50, R) >droSec1.super_4 5832379 116 - 6179234 AGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUACA----CAGAUAUCGAGUUGAGAGCGUAAAUGGGUGAACUACAUUGAAAUAAAACAAGUGCCGUGACUGCAUAU ((((((((((((((((.((((((((((((......)))))(((((...----.)))))...)))))))........)))))))))).((((...........)))).)))).))...... ( -31.80, z-score = -1.50, R) >consensus AGCACGAGUUCACUUGGCUUAAUUGGCCCAAGGACGGGCCUAUCUAC______AGAUACCGAGUUGAGAGCG_AAAUGGGUGAACUACAUUGAAAUAAAACAAGUGCUGAGACUGCAUAU ..........((((((..(((...(((((......)))))(((((........))))).....................................)))..)))))).............. (-12.06 = -12.82 + 0.76)

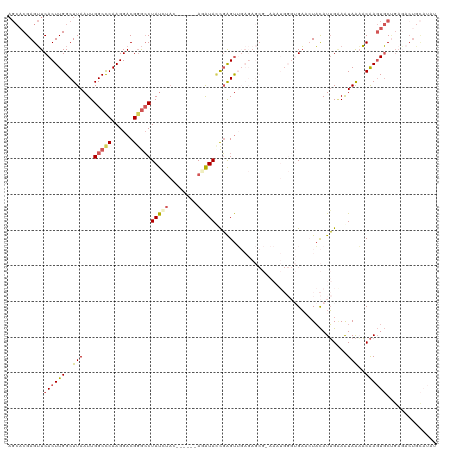
| Location | 6,445,736 – 6,445,850 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.53 |
| Shannon entropy | 0.55436 |
| G+C content | 0.44668 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.96 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
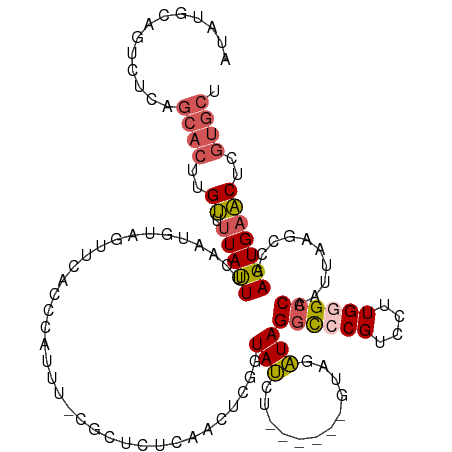
>dm3.chrX 6445736 114 - 22422827 AUAUGCAGUCUCAGCACUUGUUUUAUUCCAAUGUAGUUCACCCAUUUACGCUCUCAACUCGUUAUGU------GUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCU ............((((((((........))).(.(((((((..((((((((..............))------)))))).((((((....))))))...........))))))))))))) ( -31.84, z-score = -3.08, R) >droAna3.scaffold_12929 3012084 87 - 3277472 ---GAAAGCAUCAGAAGUCGAACUACUGCCAUAAAUUAC----------------UGCCAAGUACGU----------GUAGCUCCGUCCUUGAGCCAAUUAAGCCAAGUGAUCUCG---- ---...(((..(((.((.....)).)))..(((...(((----------------(....))))..)----------)).)))..(((((((.((.......)))))).)))....---- ( -12.70, z-score = 0.47, R) >droEre2.scaffold_4690 15385559 111 - 18748788 ---------AUAUGCACUUGUUUUACUUCAAUGUAUAUCACCCAUUUUCGCUCUCAACUCGGUAUCUGUAUAGAUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCU ---------....((((..(((((((......))).....................(((.(((((((((....)))))).((((((....))))))......))).)))))))..)))). ( -29.30, z-score = -2.68, R) >droYak2.chrX 2419834 99 - 21770863 AUAUGCAGUCUCAGCACUUGUGUUAUUUCAAUGUAGAUC-----------------ACUCGGUAUCUC----GGUAAAUAGGCGCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCU ............(((((..((.((((......)))).((-----------------(((.(((((...----.)).....(((.((....)).)))......))).)))))))..))))) ( -21.50, z-score = 0.40, R) >droSec1.super_4 5832379 116 + 6179234 AUAUGCAGUCACGGCACUUGUUUUAUUUCAAUGUAGUUCACCCAUUUACGCUCUCAACUCGAUAUCUG----UGUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCU ......((.(((((((.(((........))))))(((((((..((((((((................)----))))))).((((((....))))))...........))))))))))))) ( -32.49, z-score = -2.50, R) >consensus AUAUGCAGUCUCAGCACUUGUUUUAUUUCAAUGUAGUUCACCCAUUU_CGCUCUCAACUCGGUAUCU______GUAGAUAGGCCCGUCCUUGGGCCAAUUAAGCCAAGUGAACUCGUGCU .............((((..((.(((((...................................(((............)))((((((....))))))..........)))))))..)))). (-10.40 = -10.96 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:21:00 2011