
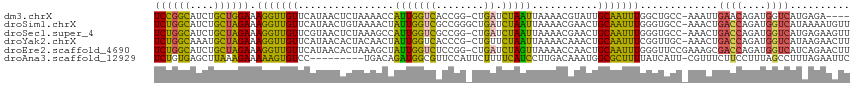
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,444,449 – 6,444,559 |
| Length | 110 |
| Max. P | 0.684660 |

| Location | 6,444,449 – 6,444,559 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.95 |
| Shannon entropy | 0.59018 |
| G+C content | 0.41363 |
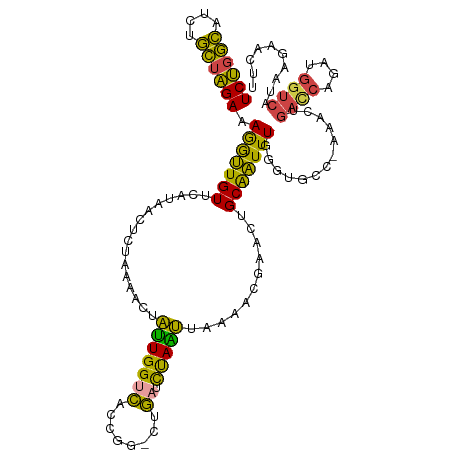
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -13.95 |
| Energy contribution | -12.26 |
| Covariance contribution | -1.68 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6444449 110 + 22422827 UCCGGCAUCUGCUGGAAAGGUUGUUCAUAACUCUAAAACCAUUGGUCACCGG-CUGAUCUAAUUAAAACGUAUUGCAAUUUGGCUGCC-AAAUUGAACAGAUGGUCAUGAGA---- ((((((....))))))..............(((....(((((((((((....-.)))))................((((((((...))-))))))....))))))...))).---- ( -28.50, z-score = -1.20, R) >droSim1.chrX 5061984 115 + 17042790 UCUGGCAUCUGCUAGAAAGGUUGUUCAUAACUGUAAAACUAUUGGUCGCCGGGCUGAUCUAAUUAAAACGAACUGCAAUUUGGGUGCC-AAACUGACCAGAUGGUCAUAAAAUGUU ((((((....))))))..(((((....))))).....(((((((((((...(((..((((((.................)))))))))-....)))))).)))))........... ( -25.73, z-score = -0.37, R) >droSec1.super_4 5831123 114 - 6179234 UCUGGCAUCUGCUAGAAAGGUUGUUCGUAACUCUAAAGCCAUUGGUCGCCGG-CUGAUCUAAUUAAAACGAACUGCAAUUUGGGUGCC-AAACUGACCAGAUGGUCAUGAGAAGUU ..((((((((....(((..((.((((((........((((..........))-))............)))))).))..))))))))))-)...(((((....)))))......... ( -29.05, z-score = -0.41, R) >droYak2.chrX 2418478 114 + 21770863 UCUGGCAAAUGCUAGAAAGGUUGUUCAUAACACUACAACUAUUGGUCACCCG-CUGUUCUAAUUAAAACAAACUGCAAUUUCGGUUGC-AAACUGACCAGAUGGUCAUAAGAACUU ((((((....))))))..((((((..........))))))...((....)).-..(((((.............((((((....)))))-)...(((((....)))))..))))).. ( -28.60, z-score = -2.51, R) >droEre2.scaffold_4690 15384247 115 + 18748788 UCUGGCAUCUGCUAGAAAGGUUGUUCAUAACACUAAAGCUAUUGGUCUCCGG-CUGAUCUAGUUAAAACCAACUGCAAUUUGGGUUCCGAAAGCGACCAGAUGGUCAUCAGAACUU ((((((....))))))......((((...........(((.((((..(((((-......(((((......)))))....)))))..)))).)))((((....))))....)))).. ( -31.40, z-score = -1.45, R) >droAna3.scaffold_12929 3009943 106 + 3277472 UCUGUGAGCUUAAAGAAAAAGUGUCC---------UGACAGAUGGCGUUCCAUUCUUUUCAUCCUUGACAAAUGGCGCUUUUAUCAUU-CGUUUCUUCCUUUAGCCUUUAGAAUUC ((((...(((.((.(((.(((((...---------(((.(((.((((..(((((....(((....)))..)))))))))))).)))..-))))).))).)).)))...)))).... ( -17.80, z-score = 0.38, R) >consensus UCUGGCAUCUGCUAGAAAGGUUGUUCAUAACUCUAAAACUAUUGGUCACCGG_CUGAUCUAAUUAAAACGAACUGCAAUUUGGGUGCC_AAACUGACCAGAUGGUCAUAAGAACUU ((((((....)))))).(((((((................(((((((........)).)))))...........))))))).............((((....)))).......... (-13.95 = -12.26 + -1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:56 2011