
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,410,494 – 6,410,594 |
| Length | 100 |
| Max. P | 0.864789 |

| Location | 6,410,494 – 6,410,594 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.43 |
| Shannon entropy | 0.47981 |
| G+C content | 0.61772 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -7.34 |
| Energy contribution | -7.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
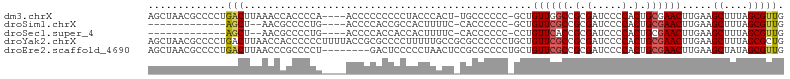
>dm3.chrX 6410494 100 + 22422827 AGCUAACGCCCCUGACUUAAACCACCCCA----ACCCCCCCCCUACCCACU-UGCCCCCC-GCUGUUGGCCGCGAUCCCCACUGCGAACUUGAAGCUUUAGCGUUG .(((((.((....................----.............(((..-.((.....-))...))).((((........))))........)).))))).... ( -14.60, z-score = -1.54, R) >droSim1.chrX 5027363 85 + 17042790 -------------AGCU--AACGCCCCUG----ACCCCACCGCCACUUUUC-CACCCCCC-GCUGUUCGCCGCGAUCCCCACUGCGAACUUGAAGCUUUAGCGUUG -------------.(((--((.((.....----........((........-........-)).((((((.(.(.....).).)))))).....)).))))).... ( -15.49, z-score = -1.97, R) >droSec1.super_4 5797409 85 - 6179234 -------------AGCU--AACGCCCCUG----ACCCCACCACCACUUUUC-CACCCCCC-CCUGUUCACCGCGAUCCCCACUGCGAACUUGAAGCUUUAGCGUUG -------------.(((--((.((...((----....))............-........-....((((.((((........))))....)))))).))))).... ( -10.40, z-score = -1.62, R) >droYak2.chrX 2382580 106 + 21770863 AGCUAACGCCCCUGACUUAACCACCCCCCUUUUACCGCGCCCCUUUUUGCCGCGCCCCCCUGCUGUUCGCCGCGAUCCCCACUGCGAACUUGAAGCUUUAGCGCUG .(((((.((...........................((((..(.....)..)))).........((((((.(.(.....).).)))))).....)).))))).... ( -22.10, z-score = -2.95, R) >droEre2.scaffold_4690 15350426 98 + 18748788 AGCUAACGCCCCUGACUUAACCCGCCCCU--------GACUCCCCCUAACUCCGCGCCCCUGCUGUUCGCCGCGAUCCCCACUGCGAACUUGAAGCUAUAGCGUUG .((....))....................--------................((((....(((((((((.(.(.....).).))))))....)))....)))).. ( -17.40, z-score = -2.00, R) >consensus AGCUAACGCCCCUGACUUAAACCCCCCCG____ACCCCACCCCCACUUACC_CGCCCCCC_GCUGUUCGCCGCGAUCCCCACUGCGAACUUGAAGCUUUAGCGUUG .............(((................................................((((((.(.(.....).).)))))).....((....))))). ( -7.34 = -7.54 + 0.20)

| Location | 6,410,494 – 6,410,594 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.43 |
| Shannon entropy | 0.47981 |
| G+C content | 0.61772 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
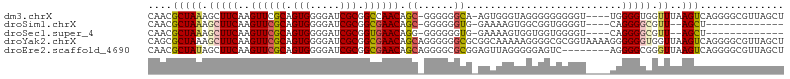
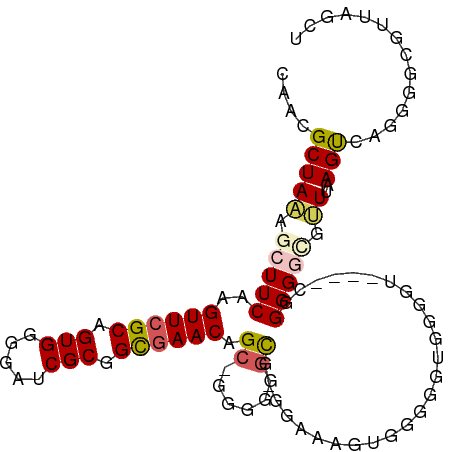
>dm3.chrX 6410494 100 - 22422827 CAACGCUAAAGCUUCAAGUUCGCAGUGGGGAUCGCGGCCAACAGC-GGGGGGCA-AGUGGGUAGGGGGGGGGU----UGGGGUGGUUUAAGUCAGGGGCGUUAGCU ....(((((.(((((..(..(((.(((.....))).(((..(...-.)..))).-.)))..).......((.(----((((....))))).)).))))).))))). ( -26.10, z-score = -0.08, R) >droSim1.chrX 5027363 85 - 17042790 CAACGCUAAAGCUUCAAGUUCGCAGUGGGGAUCGCGGCGAACAGC-GGGGGGUG-GAAAAGUGGCGGUGGGGU----CAGGGGCGUU--AGCU------------- ....(((((.(((((..((((((.(((.....))).))))))...-........-......((((......))----))))))).))--))).------------- ( -28.00, z-score = -1.90, R) >droSec1.super_4 5797409 85 + 6179234 CAACGCUAAAGCUUCAAGUUCGCAGUGGGGAUCGCGGUGAACAGG-GGGGGGUG-GAAAAGUGGUGGUGGGGU----CAGGGGCGUU--AGCU------------- ....(((((.(((((..((((((.(((.....))).))))))...-........-.........(((.....)----))))))).))--))).------------- ( -22.20, z-score = -1.13, R) >droYak2.chrX 2382580 106 - 21770863 CAGCGCUAAAGCUUCAAGUUCGCAGUGGGGAUCGCGGCGAACAGCAGGGGGGCGCGGCAAAAAGGGGCGCGGUAAAAGGGGGGUGGUUAAGUCAGGGGCGUUAGCU ....(((((.(((((..((((((.(((.....))).))))))........(.((((.(.......).)))).).....................))))).))))). ( -32.60, z-score = -1.47, R) >droEre2.scaffold_4690 15350426 98 - 18748788 CAACGCUAUAGCUUCAAGUUCGCAGUGGGGAUCGCGGCGAACAGCAGGGGCGCGGAGUUAGGGGGAGUC--------AGGGGCGGGUUAAGUCAGGGGCGUUAGCU ....(((((((((((..((((((.(((.....))).)))))).((......)))))))))......(((--------...(((.......)))...)))..)))). ( -31.40, z-score = -2.22, R) >consensus CAACGCUAAAGCUUCAAGUUCGCAGUGGGGAUCGCGGCGAACAGC_GGGGGGCG_GGAAAGUGGGGGUGGGGU____CGGGGGCGUUUAAGUCAGGGGCGUUAGCU ....(((...(((((..((((((.(((.....))).)))))).((......))..........................))))).....))).............. (-14.22 = -14.70 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:52 2011