| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,397,743 – 6,397,811 |
| Length | 68 |
| Max. P | 0.654832 |

| Location | 6,397,743 – 6,397,811 |
|---|---|
| Length | 68 |
| Sequences | 9 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.38950 |
| G+C content | 0.43485 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.46 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
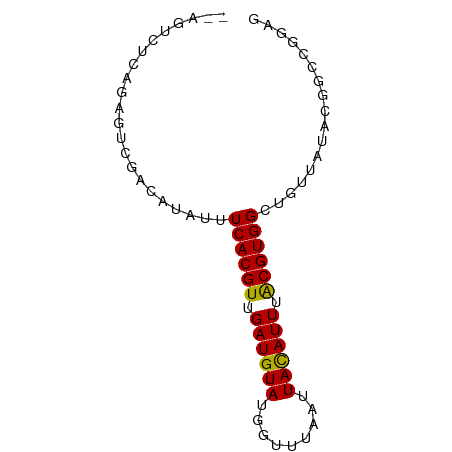
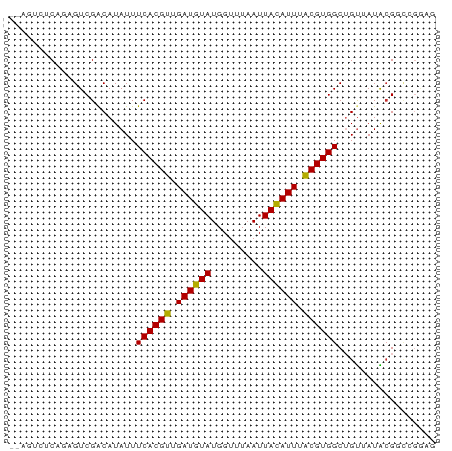
>dm3.chrX 6397743 68 - 22422827 ------UCAGUGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUACGGCCGAAG ------...(((....)))..(((((((.((((((.........)))))).))))(((((.....)))))))). ( -18.00, z-score = -1.79, R) >droPer1.super_18 1817008 71 - 1952607 --GUAGUCGACGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUGGGCCGGGG- --...((((....))))...(..(((((.((((((.........)))))).))))((((......)))))..)- ( -19.80, z-score = -1.52, R) >droEre2.scaffold_4690 15338004 74 - 18748788 GCAGUCGCAGAGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUACGGCCGGCG ((.((((......)))).......((((.((((((.........)))))).))))(((((.....))))).)). ( -22.30, z-score = -1.87, R) >droYak2.chrX 2369580 74 - 21770863 GCAGUCGCAGAGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUACGGCCGGAG ...((((......))))....(((((((.((((((.........)))))).))))(((((.....)))))))). ( -21.20, z-score = -1.91, R) >droSec1.super_4 5784902 74 + 6179234 GCAGUCGCAGAGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUAUAUUUACGUGGCUGUUAUACGGCCGGAG ...((((......))))....(((((((.((((((.........)))))).))))(((((.....)))))))). ( -19.30, z-score = -1.27, R) >droSim1.chrX 5014711 74 - 17042790 GCAGUCGCAGAGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUACGGCCGGAG ...((((......))))....(((((((.((((((.........)))))).))))(((((.....)))))))). ( -21.20, z-score = -1.91, R) >droVir3.scaffold_12928 7695159 68 - 7717345 ------UCGCCGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUGCGUGGCGGCAAUGCGUUUGGGA ------..((((((((......))((((.((((((.........)))))).))))))))))............. ( -20.00, z-score = -1.05, R) >droGri2.scaffold_14853 669511 61 + 10151454 ------UCACCGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUGCGUGGCAGCAAUGCG------- ------.................(((((.((((((.........)))))).)))))(((....))).------- ( -15.80, z-score = -1.24, R) >droWil1.scaffold_181150 140677 65 + 4952429 --GUGCUCGUUGUCGGCAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUAAUACG------- --((((.((....))))))...((((((.((((((.........)))))).))))))..........------- ( -16.50, z-score = -1.50, R) >consensus __AGUCUCAGAGUCGACAUAUUUCACGUUGAUGUAUGGUUUAAUUACAUUUACGUGGCUGUUAUACGGCCGGAG ......................((((((.((((((.........)))))).))))))................. (-11.73 = -11.46 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:50 2011