| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,395,224 – 6,395,352 |
| Length | 128 |
| Max. P | 0.862264 |

| Location | 6,395,224 – 6,395,332 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.97 |
| Shannon entropy | 0.30008 |
| G+C content | 0.52323 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862264 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
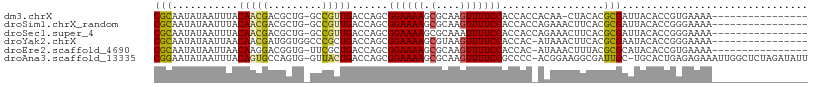
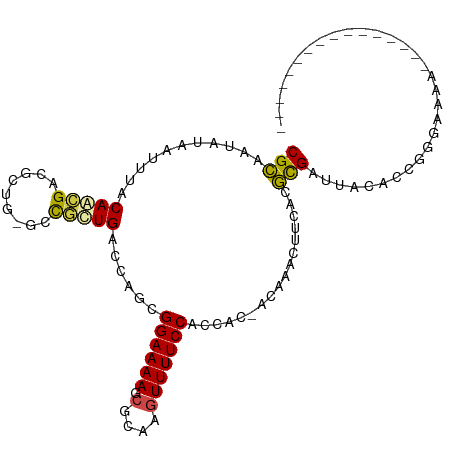
>dm3.chrX 6395224 108 + 22422827 ---GGACGGACAGAGACAGUGACAGGCAACUGACAACCGGCGCAAUAUAAUUUACAACGACGCUGGC-CGUUGACCAGCGGAAAAGCGCAAGUUUUCCACCACCACAA-CUAC ---((((((..........((.(((....))).)).(((((((...............).)))))))-)))........((((((.(....))))))).)).......-.... ( -27.96, z-score = -1.94, R) >droSim1.chrX_random 2077756 109 + 5698898 ---GGACGGACAGAGACAGUGACAGGCAACUGACACCCGGCGCAAUAUAAUUUACAACGACGCUGGC-CGUUGACCAGCGGAAAAGCGCAAGUUUUCCACCACCAGAAACUUC ---((((((.........(((.(((....))).)))(((((((...............).)))))))-)))........((((((.(....))))))).))............ ( -30.46, z-score = -2.22, R) >droSec1.super_4 5782493 109 - 6179234 ---GGACGGACAGAGACAGUGACAGGCAACUGACACCCGGCGCAAUAUAAUUUACAACGACGCUGGC-CGUUGACCAGCGGAAAAGCGCAAAUUUUCCACCACCAGAAACUUC ---((((((.........(((.(((....))).))))))((((((......)).......((((((.-......)))))).....))))......)))............... ( -28.30, z-score = -2.01, R) >droYak2.chrX 2367165 109 + 21770863 -AAGAGCGGACGGA--CAGUGACAGGCAACUGACACCCGGCGCAAUAUAAUUAACAACGAUGGUGGCCCGCUGACCAGCGGAAAAGCGUAAGUUUUCCACCAC-AUAAACUUC -....(((..(((.--..(((.(((....))).)))))).)))...............(.((((((.(((((....))))).(((((....))))))))))))-......... ( -37.30, z-score = -3.75, R) >droEre2.scaffold_4690 15335637 106 + 18748788 ---GGACGGACGGA--CAGUGACAGGCAACUGACACCCGGCGCAAUAUAAUUAACAAGGACGGUG-UUCGCUGACCAGCGGAAAAGCGCAAGUUUUCCACCAC-AUAAACUUU ---((.(((.((((--(((((.(((....))).)))(((.....................)))))-))))))).))...((((((.(....))))))).....-......... ( -30.80, z-score = -2.18, R) >droAna3.scaffold_13335 1367947 105 - 3335858 CAUGGAGAGGCUUA--CAGUGACAGGCAACUGACACCCGGCGGAAUAUAAUUUACAGUGCCAGUGGU-UACUGACCAGCGGAAAAGCGCAAGUUUUCCGCCCCACGGA----- ..(((...((....--(((((((((....))..(((..((((...((.....))...)))).)))))-))))).)).((((((((.(....))))))))).)))....----- ( -33.40, z-score = -1.59, R) >consensus ___GGACGGACAGA__CAGUGACAGGCAACUGACACCCGGCGCAAUAUAAUUUACAACGACGCUGGC_CGCUGACCAGCGGAAAAGCGCAAGUUUUCCACCACCAGAAACUUC ...(((.((((.......(((.(((....))).)))...((((...........(((((.........))))).((...))....))))..)))))))............... (-19.38 = -19.47 + 0.09)
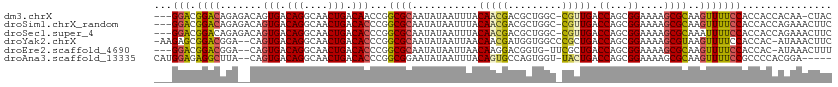
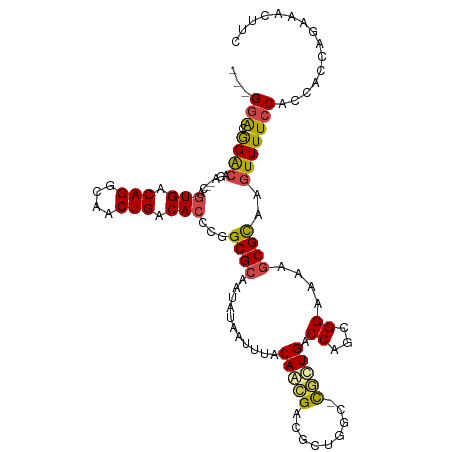
| Location | 6,395,261 – 6,395,352 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Shannon entropy | 0.37306 |
| G+C content | 0.47500 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6395261 91 + 22422827 CGCAAUAUAAUUUACAACGACGCUG-GCCGUUGACCAGCGGAAAAGCGCAAGUUUUCCACCACCACAA-CUACACGCGAUUACACCGUGAAAA---------------- (((..................((((-(.......)))))((((((.(....)))))))..........-......)))...............---------------- ( -20.80, z-score = -1.83, R) >droSim1.chrX_random 2077793 92 + 5698898 CGCAAUAUAAUUUACAACGACGCUG-GCCGUUGACCAGCGGAAAAGCGCAAGUUUUCCACCACCAGAAACUUCACGCGAUUACACCGGGAAAA---------------- (((..................((((-(.......)))))((((((.(....))))))).................)))...............---------------- ( -20.80, z-score = -1.12, R) >droSec1.super_4 5782530 92 - 6179234 CGCAAUAUAAUUUACAACGACGCUG-GCCGUUGACCAGCGGAAAAGCGCAAAUUUUCCACCACCAGAAACUUCACGCGAUUACACCGGGAAAA---------------- (((..................((((-(.......)))))((((((.......)))))).................)))...............---------------- ( -18.00, z-score = -0.58, R) >droYak2.chrX 2367202 92 + 21770863 CGCAAUAUAAUUAACAACGAUGGUGGCCCGCUGACCAGCGGAAAAGCGUAAGUUUUCCACCAC-AUAAACUUCACGCGAAUACACCGGGAAAA---------------- ....................(((((..(((((....)))))....((((((((((........-..)))))).)))).....)))))......---------------- ( -23.70, z-score = -1.96, R) >droEre2.scaffold_4690 15335672 91 + 18748788 CGCAAUAUAAUUAACAAGGACGGUG-UUCGCUGACCAGCGGAAAAGCGCAAGUUUUCCACCAC-AUAAACUUUACGCGCAUACACCGUGAAAA---------------- ...................((((((-((((((....))))))...((((((((((........-..))))))...))))...)))))).....---------------- ( -24.80, z-score = -2.63, R) >droAna3.scaffold_13335 1367985 106 - 3335858 CGGAAUAUAAUUUACAGUGCCAGUG-GUUACUGACCAGCGGAAAAGCGCAAGUUUUCCGCCCC-ACGGAAGGCGAUUGC-UGCACUGAGAGAAAUUGGCUCUAGAUAUU ...(((((......((((((.((..-((..((..((.((((((((.(....)))))))))...-..))..))..))..)-)))))))((((.......))))..))))) ( -35.50, z-score = -2.90, R) >consensus CGCAAUAUAAUUUACAACGACGCUG_GCCGCUGACCAGCGGAAAAGCGCAAGUUUUCCACCAC_ACAAACUUCACGCGAUUACACCGGGAAAA________________ (((...........(((((.........)))))......((((((.(....))))))).................)))............................... (-12.84 = -12.90 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:49 2011