
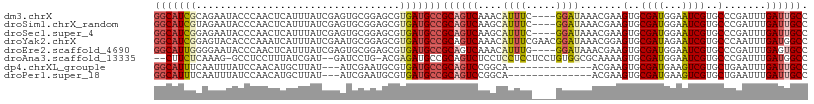
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,387,229 – 6,387,365 |
| Length | 136 |
| Max. P | 0.859821 |

| Location | 6,387,229 – 6,387,334 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.08 |
| Shannon entropy | 0.53550 |
| G+C content | 0.50271 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -14.73 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
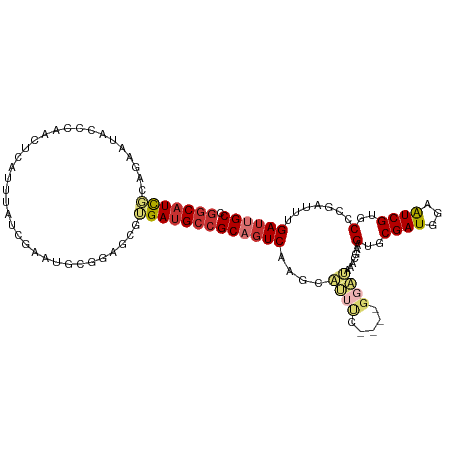
>dm3.chrX 6387229 105 + 22422827 GGCAUCGCAGAAUACCCAACUCAUUUAUCGAGUGCGGAGCGUGAUGCCGCAGUCAAACAUUUC----GGAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCC ((((((((.(.....((.((((.......))))..))..)))))))))((((((...((((((----(......))))))).....((((((....)))))))))))). ( -38.00, z-score = -2.94, R) >droSim1.chrX_random 2070015 105 + 5698898 GGCAUCGUAGAAUACCCAACUCAUUUAUCGAGUGCGGAGCGUGAUGCCGCAGUCAAGCAUUUC----GGAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCC (((((((((...)))((.((((.......))))..)).....))))))((((((..(((((((----(......))))))))....((((((....)))))))))))). ( -39.40, z-score = -3.33, R) >droSec1.super_4 5774617 105 - 6179234 GGCAUCGGAGAAUACCCAACUCAUUUAUCGAGUGCGGAGCGUGAUGCCGCAGUCAAGCAUUUC----GGAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCC (((((((..(.....((.((((.......))))..))..).)))))))((((((..(((((((----(......))))))))....((((((....)))))))))))). ( -39.70, z-score = -3.07, R) >droYak2.chrX 2359163 109 + 21770863 GGCAUCGGAGUACACCCAAAUCAUUUAUCGAAUGCGGAGCGUGAUGCCGCAGUCAAACAUUUCGAACGGAUAAACGGAGUGCGAUAGAAUCGUGCCCAAUUUGAUGGCC (((...((.(....)))........((((((((..((.(((((((..((((.((......(((....)))......)).)))).....))))))))).))))))))))) ( -29.40, z-score = -0.18, R) >droEre2.scaffold_4690 15327742 105 + 18748788 GGCAUUGGGGAAUACCCAACUCAUUUAUCGAGUGCGGAGCGUGAUGCCGCAGUCAAACAUUUG----GGAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAGUGCC (((..((((.....))))((((...((((((.(((((.........))))).))...(((((.----(......).))))).))))((((((....))))))))))))) ( -35.40, z-score = -1.69, R) >droAna3.scaffold_13335 1358991 103 - 3335858 --CUUCUCAAAG-GCCUCCUUUAUCGAU--GAUCCUG-ACGAGAUGCCGCAGUCUCCUCCUCCUCCUGUGGCGCAAAAGUGCGAUGGAAUCGUGCCCGAUUUGAUGGCC --.........(-(((..(...((((((--(((((..-.......(((((((.............)))))))(((....)))...)).)))))...))))..)..)))) ( -28.22, z-score = -0.59, R) >dp4.chrXL_group1e 2911264 92 + 12523060 GGCAUUUCAAUUUAUCCAACAUGCUUAU---AUCGAAUGCGUGAUGCCGCAGUCCGGCA--------------ACGAAGUGCGAUGAAGUCGUGCUGAAUUUGAUUGCC (((((..............(((((....---.......))))))))))((((((((...--------------.)).((..((((...))))..))......)))))). ( -27.33, z-score = -1.21, R) >droPer1.super_18 1806292 92 + 1952607 GGCAUUUCAAUUUAUCCAACAUGCUUAU---AUCGAAUGCGUGAUGCCGCAGUCCGGCA--------------ACGAAGUGCGAUGAAGUCGUGCUGAAUUUGAUUGCC (((((..............(((((....---.......))))))))))((((((((...--------------.)).((..((((...))))..))......)))))). ( -27.33, z-score = -1.21, R) >consensus GGCAUCGCAGAAUACCCAACUCAUUUAUCGAAUGCGGAGCGUGAUGCCGCAGUCAAGCAUUUC____GGAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCC (((((((..................................)))))))((((((....((((.....)))).......(..((((...))))..).......)))))). (-14.73 = -15.33 + 0.60)
| Location | 6,387,265 – 6,387,365 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Shannon entropy | 0.40830 |
| G+C content | 0.53475 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
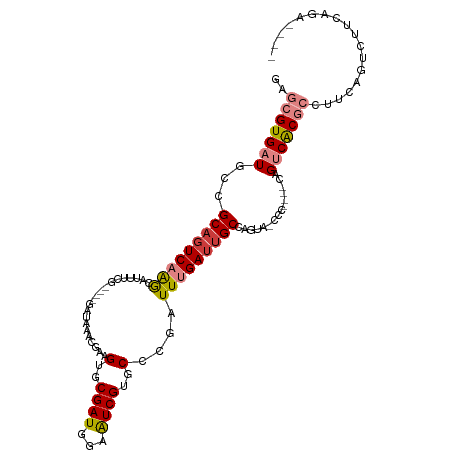
>dm3.chrX 6387265 100 + 22422827 GAGCGUGAUGCCGCAGUCAAACAUUUCG----GAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCCAAUAGCCC----CAGUCACGCCUUCAGUCUUCAGA---- ..(((((((...((........((((((----......))))))(((((.((((((....)))))).))))).....))..----..)))))))..............---- ( -30.40, z-score = -2.02, R) >droSim1.chrX_random 2070051 99 + 5698898 GAGCGUGAUGCCGCAGUCAAGCAUUUCG----GAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCCAAUA-CCC----CAGUCACGCCUUCAGUCUUCAGA---- ..(((((((...((((((..((((((((----......))))))))....((((((....)))))))))))).....-...----..)))))))..............---- ( -32.72, z-score = -2.75, R) >droSec1.super_4 5774653 99 - 6179234 GAGCGUGAUGCCGCAGUCAAGCAUUUCG----GAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCCAAUA-CCC----CAGUCACGCAUUCAGUCUUCAGA---- ..(((((((...((((((..((((((((----......))))))))....((((((....)))))))))))).....-...----..)))))))..............---- ( -33.62, z-score = -2.98, R) >droYak2.chrX 2359199 112 + 21770863 GAGCGUGAUGCCGCAGUCAAACAUUUCGAACGGAUAAACGGAGUGCGAUAGAAUCGUGCCCAAUUUGAUGGCCAGUACCCCACGCCAGUCACGCCUUCCGUCUUCAGACUAC ..(((((((...((.((((((...(((....))).....((.(..((((...))))..)))..))))))((........))..))..))))))).....(((....)))... ( -28.90, z-score = -0.52, R) >droEre2.scaffold_4690 15327778 103 + 18748788 GAGCGUGAUGCCGCAGUCAAACAUUUGG----GAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAGUGCCAGUA-CCC----CAGUCACGCCUUCAGUUUUCAGACUAC ..(((((((((.(((.(((((...((((----(....((((............)))).)))))))))).)))..)).-...----..)))))))....((((....)))).. ( -27.00, z-score = -0.54, R) >dp4.chrXL_group1e 2911300 86 + 12523060 ---CGUGAUGCCGCAGUCCGGCA--------------ACGAAGUGCGAUGAAGUCGUGCUGAAUUUGAUUGCCAGU--CCCGACCCAGGCGCACGACC---CUUCAAA---- ---((((.((((((((((((...--------------.)).((..((((...))))..))......))))))..((--....))...))))))))...---.......---- ( -28.50, z-score = -1.73, R) >droPer1.super_18 1806328 86 + 1952607 ---CGUGAUGCCGCAGUCCGGCA--------------ACGAAGUGCGAUGAAGUCGUGCUGAAUUUGAUUGCCAGU--CCCCACCCAGGCGCACGACC---CUUCAAA---- ---((((.((((((((((((...--------------.)).((..((((...))))..))......))))))..((--....))...))))))))...---.......---- ( -27.40, z-score = -1.64, R) >consensus GAGCGUGAUGCCGCAGUCAAGCAUUUCG____GAUAAACGAAGUGCGAUGGAAUCGUGCCCGAUUUGAUUGCCAGUA_CCC____CAGUCACGCCUUCAGUCUUCAGA____ ..(((((((...(((((((((....((.....))........(..((((...))))..)....)))))))))...............))))))).................. (-15.18 = -16.04 + 0.86)

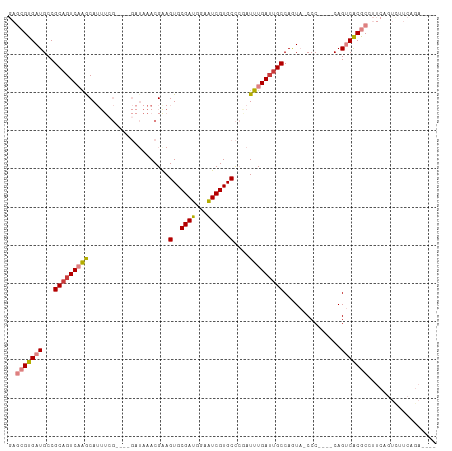
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:46 2011