
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,357,912 – 6,358,028 |
| Length | 116 |
| Max. P | 0.682790 |

| Location | 6,357,912 – 6,358,028 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.56968 |
| G+C content | 0.37352 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -13.51 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
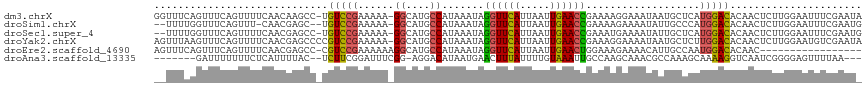
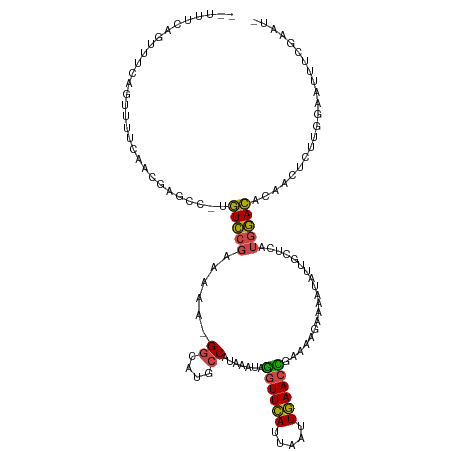
>dm3.chrX 6357912 116 + 22422827 GGUUUCAGUUUCAGUUUUCAACAAGCC-UGUCCGAAAAA-GGCAUGCCAUAAAUAGGUUCAUUAAUUGAACCGAAAAGGAAAUAAUGCUCAUGGACACAACUCUUGGAAUUUCGAAUA ((((((((((((..(((((.....(((-(.........)-)))............((((((.....))))))))))).)))))........((....)).....)))))))....... ( -23.70, z-score = -0.56, R) >droSim1.chrX 5002079 112 + 17042790 --UUUUGGUUUCAGUUU-CAACGAGC--UGUCCGAAAAA-GGCAUGCCAUAAAUAGGUUCAUUAAUUGAACCGAAAAGAAAAUAUUGCCCAUGGACACAACUCUUGGAAUUUCGAAUG --.(((((....(((((-(((.(((.--((((((.....-((((...........((((((.....)))))).............))))..))))))...)))))))))))))))).. ( -27.75, z-score = -2.05, R) >droSec1.super_4 5748107 114 - 6179234 --UUUUGGUUUCAGUUUUCAACGAGCC-UGUCCGAAAAA-GGCAUGCCAUAAAUAGGUUCAUUAAUUGAACCGAAAUGAAAAUAUUGCUCAUGGACACAACUCUUGGAAUUUCGAAUG --.....((((.((.((((((.(((..-((((((.....-((....)).......((((((.....))))))...................))))))...))))))))).)).)))). ( -25.60, z-score = -1.19, R) >droYak2.chrX 2334021 117 + 21770863 AGUUUAAGUUUCAGUUUUCAACGAGCCCCGUCCGAAAAA-GGCAUGCCAUAAAUAGGUUCAUUAAUUGAACCGAAAGGAAAAUAAUGCUCUUGGACACAACUCUUGGAAUGUCGAAUA .......((((....((((((.(((....((((((....-(((((..........((((((.....))))))............))))).))))))....)))))))))....)))). ( -26.45, z-score = -1.19, R) >droEre2.scaffold_4690 15302474 100 + 18748788 AGUUUCAGUUUCAGUUUUCAACGAGCC-CGUCCGAAAAAAGGCAUGCCAUAAAUAGGUUCAUUAAUUGAACUGGAAAGAAAACAUUGCCAAUGGACACAAC----------------- .((((((....((((((((..((....-))(((.......((....)).......((((((.....)))))))))..))))))..))....)))).))...----------------- ( -18.00, z-score = -0.15, R) >droAna3.scaffold_13335 1325476 105 - 3335858 -------GAUUUUUUUCUCAUUUUAC--UCUUCGGAUUUCGG-AGGACAUAAUGAACUUUAUUUUGUAAAUUGCCAAGCAAACGCCAAAGCAAAAGGUCAAUCGGGGAGUUUUAA--- -------......((((((.......--(((((.(....).)-)))).....(((.((((.(((((....((((...))))....)))))..)))).)))...))))))......--- ( -16.70, z-score = 0.61, R) >consensus __UUUCAGUUUCAGUUUUCAACGAGCC_UGUCCGAAAAA_GGCAUGCCAUAAAUAGGUUCAUUAAUUGAACCGAAAAGAAAAUAUUGCUCAUGGACACAACUCUUGGAAUUUCGAAU_ .............................(((((......((....)).......((((((.....))))))...................)))))...................... (-13.51 = -12.98 + -0.52)
| Location | 6,357,912 – 6,358,028 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.56968 |
| G+C content | 0.37352 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
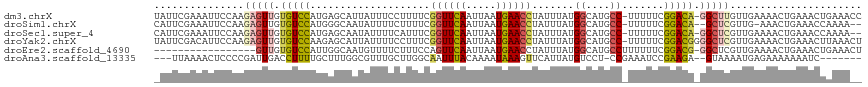
>dm3.chrX 6357912 116 - 22422827 UAUUCGAAAUUCCAAGAGUUGUGUCCAUGAGCAUUAUUUCCUUUUCGGUUCAAUUAAUGAACCUAUUUAUGGCAUGCC-UUUUUCGGACA-GGCUUGUUGAAAACUGAAACUGAAACC ..((((...(((...(((((.(((((....................((((((.....)))))).......((....))-......)))))-)))))...)))...))))......... ( -22.60, z-score = -0.24, R) >droSim1.chrX 5002079 112 - 17042790 CAUUCGAAAUUCCAAGAGUUGUGUCCAUGGGCAAUAUUUUCUUUUCGGUUCAAUUAAUGAACCUAUUUAUGGCAUGCC-UUUUUCGGACA--GCUCGUUG-AAACUGAAACCAAAA-- ..((((...(((...((((..(((((..(((((.............((((((.....))))))...........))))-).....)))))--))))...)-))..)))).......-- ( -24.85, z-score = -1.00, R) >droSec1.super_4 5748107 114 + 6179234 CAUUCGAAAUUCCAAGAGUUGUGUCCAUGAGCAAUAUUUUCAUUUCGGUUCAAUUAAUGAACCUAUUUAUGGCAUGCC-UUUUUCGGACA-GGCUCGUUGAAAACUGAAACCAAAA-- ..((((...(((...(((((.((((((((((.......)))))...((((((.....)))))).......((....))-......)))))-)))))...)))...)))).......-- ( -24.40, z-score = -0.88, R) >droYak2.chrX 2334021 117 - 21770863 UAUUCGACAUUCCAAGAGUUGUGUCCAAGAGCAUUAUUUUCCUUUCGGUUCAAUUAAUGAACCUAUUUAUGGCAUGCC-UUUUUCGGACGGGGCUCGUUGAAAACUGAAACUUAAACU ..((((...(((...(((((.(((((....................((((((.....)))))).......((....))-......))))).)))))...)))...))))......... ( -27.30, z-score = -1.28, R) >droEre2.scaffold_4690 15302474 100 - 18748788 -----------------GUUGUGUCCAUUGGCAAUGUUUUCUUUCCAGUUCAAUUAAUGAACCUAUUUAUGGCAUGCCUUUUUUCGGACG-GGCUCGUUGAAAACUGAAACUGAAACU -----------------....((((....))))..(((((..((.(((((...(((((((.((.......))...((((.((....)).)-)))))))))).))))).))..))))). ( -18.20, z-score = 0.37, R) >droAna3.scaffold_13335 1325476 105 + 3335858 ---UUAAAACUCCCCGAUUGACCUUUUGCUUUGGCGUUUGCUUGGCAAUUUACAAAAUAAAGUUCAUUAUGUCCU-CCGAAAUCCGAAGA--GUAAAAUGAGAAAAAAAUC------- ---............((((......(((((..(((....))).)))))..............((((((.((..((-.((.....)).)).--.)).)))))).....))))------- ( -14.50, z-score = 0.25, R) >consensus _AUUCGAAAUUCCAAGAGUUGUGUCCAUGAGCAAUAUUUUCUUUUCGGUUCAAUUAAUGAACCUAUUUAUGGCAUGCC_UUUUUCGGACA_GGCUCGUUGAAAACUGAAACUAAAA__ ...............(((((.(((((....................((((((.....)))))).......((....)).......))))).)))))...................... (-14.47 = -14.87 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:40 2011