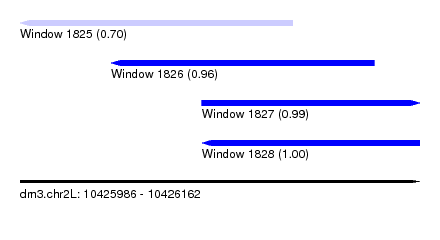
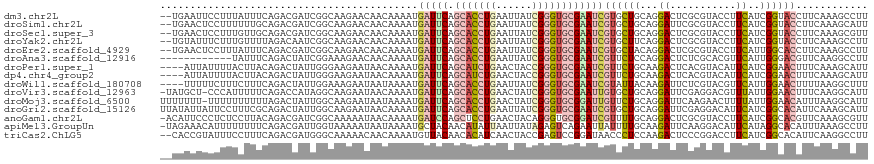
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,425,986 – 10,426,162 |
| Length | 176 |
| Max. P | 0.999726 |

| Location | 10,425,986 – 10,426,106 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Shannon entropy | 0.39434 |
| G+C content | 0.45333 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.35 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10425986 120 - 23011544 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCCUUCGACAAACACAUGAACUUGAUCCUCGGCGACUGCGAGGAG ...(((((((.((((((((....))))))))))))))).........((((((((((......))))).......(((...(((((..........))).))...)))....)))))... ( -40.00, z-score = -2.75, R) >droSim1.chr2L 10217775 120 - 22036055 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGAUUCGCGUACCUUCAUCGGUACCUUCAAAGCAUUCGACAAACACAUGAACUUGAUCCUCGGCGACUGCGAGGAG .....(((((.((((((((....)))))))))))))(((((....((...(.(((((......)))))).))..))))).....................((((((.......)))))). ( -39.00, z-score = -2.47, R) >droSec1.super_3 5853886 120 - 7220098 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCGUUCGACAAACACAUGAACUUGAUCCUCGGCGACUGCGAGGAG ...(((((((.((((((((....)))))))))))))))(((((.......))(((((......)))))......)))(((((.........)))))....((((((.......)))))). ( -40.70, z-score = -2.59, R) >droYak2.chr2L 13106512 120 - 22324452 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUUCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCCUUCGACAAACACAUGAACUUGAUCCUAGGAGAUUGCGAGGAG ...(((((((.((((((((....))))))))))))))).........((((((((((......)))))........(((..(((((..........))).))..))).....)))))... ( -38.70, z-score = -2.91, R) >droEre2.scaffold_4929 11035287 120 - 26641161 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUACAGGACUCGCGUACCUUCAUUGGCACCUUCAAAGCCUUUGACAAACACAUGAACCUGAUCCUCGGCGAUUGCGAGGAG ...(((((((.((((((((....)))))))))))))))....((((..(((.((....(((..(((.........)))..))).....)).))).)))).((((((.......)))))). ( -41.90, z-score = -3.96, R) >droAna3.scaffold_12916 14267682 120 - 16180835 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUUCUCCAGGACUCUCGCACGUUCAUUGGGACGUUCAAGGCCUUCGACAAACACAUGAAUCUUAUUCUCGGAGACUGCGAGGAG .....(((((.((((((((....)))))))))))))((((....))))((((((((((((....))))))....((.((((((....................)))))).)))))))).. ( -40.75, z-score = -2.98, R) >droPer1.super_1 9293909 120 + 10282868 AAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUCUGCAAGACUCACGUACAUUCAUCGGAACUUUCAAAGCAUUUGAUAAGCACAUGAACUUAAUAUUGGGCGACUGCGAAGAG ..((((((((.(((((((......))))))))))))))).((((.(.((((..((..(((((.(...(((((((.....)))).)))..))))))..))....)))))...))))..... ( -26.90, z-score = -0.59, R) >dp4.chr4_group2 192456 120 + 1235136 AAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUCUGCAAGACUCACGUACAUUCAUCGGAACUUUCAAAGCAUUUGAUAAGCACAUGAACUUAAUAUUGGGCGACUGCGAAGAG ..((((((((.(((((((......))))))))))))))).((((.(.((((..((..(((((.(...(((((((.....)))).)))..))))))..))....)))))...))))..... ( -26.90, z-score = -0.59, R) >droWil1.scaffold_180708 4863863 120 - 12563649 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUAUUACAAGAUUCUCGUACGUUCAUUGGAACUUUUAAGGCUUUCGACAAGCAUAUGAACCUCAUAUUGGGUGACUGUGAGGAA ...(((((((.((((((((....)))))))))))))))............(((((.(((..((((((((.....)).))))))..))).))))).((((((...((....)))))))).. ( -39.60, z-score = -4.03, R) >droVir3.scaffold_12963 19183933 120 + 20206255 AAAAUGAUUCAGCACCUGAACUAUCGGGUGCGAAUUGUGCUGCAGGAUUCGAGGACGUUUAUUGGAACUUUCAAGGCAUUUGAUAAGCAUAUGAAUCUUAUAUUGGGCGAUUGCGAAGAG .......(((.((((((((....)))))))).((((((.(...((((((((.(...((((((..((.((....))...))..)))))).).)))))))).....).))))))..)))... ( -32.30, z-score = -1.58, R) >droMoj3.scaffold_6500 4600080 120 + 32352404 AAAAUGAUUCAGCACCUGAACUAUCGGGUGCGGAUUGUUCUGCAGGAUUCAAGAACUUUUAUUGGAACAUUUAAGGCAUUCGAUAAGCAUAUGAAUCUUAUAUUGGGCGAUUGCGAAGAG .......(((.((((((((....)))))))).((((((((...((((((((......(((((((((.(......)...)))))))))....)))))))).....))))))))..)))... ( -31.60, z-score = -2.03, R) >droGri2.scaffold_15126 2426105 120 - 8399593 AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGAUUCGAGGACAUUCAUCGGCACAUUCAAAGCAUUCGAUAAGCACAUGAAUCUGAUAUUGGGCGACUGCGAGGAG .....(((((.((((((((....)))))))))))))((((((..((((........))))..))))))..((...(((.(((.(((.((........))...)))..))).)))...)). ( -36.40, z-score = -1.69, R) >anoGam1.chr2L 32754120 120 + 48795086 AAAAUGAUCCAGCUCCUGAACUACAGGGUGCGGAUCGUUUUGCAGGACUCGCGUACCUUCAUCGGCACGUUCAAAGCGUUCGACAAACACAUGAACCUCAUCCUGGGGGACUGCGAGGAG ((((((((((.(((((((.....))))).))))))))))))......(((((((.((((((((((.((((.....)))))))).......(((.....)))..)))))))).)))))... ( -42.10, z-score = -2.28, R) >apiMel3.GroupUn 18893672 120 + 399230636 AAAAUGCUACAACAUAUUAAUUAUAGAGUCAGAAUUAUUUUGCAAGAUUCAAGGACAUUCAUAGGCACAUUUAAAGCCUUUGAUAAACAUAUGAAUCUUAUACUUGGUGAUUGUGAAGAA ............................(((.(((((((..(.((((((((.......(((.((((.........)))).)))........))))))))...)..))))))).))).... ( -22.56, z-score = -1.23, R) >triCas2.ChLG5 9637761 120 - 18847211 AAAAUGUUACAACACAUCAACUACCGAGUCCGGAUAACCCUCCAAGACUCCCGGACCUUCAUCGGCACAUUCAAGGCCUUCGACAAACACAUGAACAUGAUCCUCGGCGACUGCGAGGAA ...(((((.((............(((((((.(((......)))..)))))..)).........(((.(......)))).............)))))))..((((((.......)))))). ( -25.50, z-score = -0.58, R) >consensus AAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGAACCUUCAAAGCAUUCGACAAACACAUGAACCUGAUACUGGGCGACUGCGAGGAG .....(((((.(((((((......))))))))))))((((((...((...........))..)))))).(((...((..(((.........................)))..))...))) (-16.47 = -16.35 + -0.12)
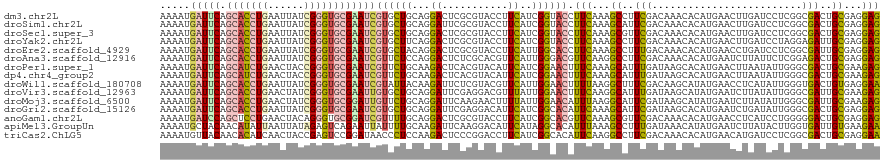
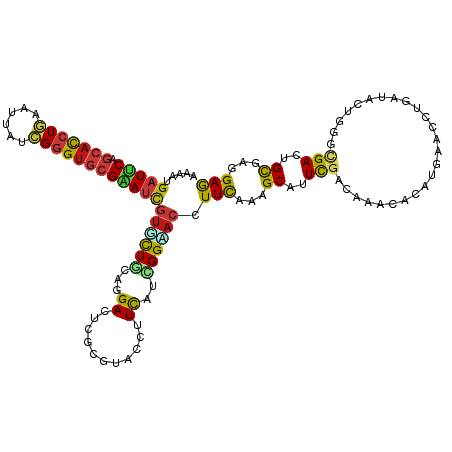
| Location | 10,426,026 – 10,426,142 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.51890 |
| G+C content | 0.41312 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10426026 116 - 23011544 --UGAAUUCCUUUAUUUCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCCUU --((((.........)))).......(((.((..(..((..(((((((.((((((((....)))))))))))))))..))..)..))...(((((......))))).......))).. ( -33.70, z-score = -2.39, R) >droSim1.chr2L 10217815 116 - 22036055 --UGAACUCCUUUUUUGCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGAUUCGCGUACCUUCAUCGGUACCUUCAAAGCAUU --.(((.(((((((((((.(.....).)))))))...((..(((((((.((((((((....)))))))))))))))..)).)))))))..(((((......)))))............ ( -38.20, z-score = -3.38, R) >droSec1.super_3 5853926 116 - 7220098 --UGAACUCCUUUGUUGCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCGUU --..(((..(((((((((((.((.((.....)).)......(((((((.((((((((....)))))))))))))))))))))).......(((((......)))))...))))).))) ( -37.00, z-score = -2.33, R) >droYak2.chr2L 13106552 116 - 22324452 --UGUAUUUCUUUGUUUUAGACAAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUUCAGGACUCGCGUACCUUCAUCGGUACCUUCAAAGCCUU --.......(((((....((.......((.((..(......(((((((.((((((((....)))))))))))))))......)..)).))(((((......))))))).))))).... ( -33.00, z-score = -2.73, R) >droEre2.scaffold_4929 11035327 116 - 26641161 --UGAACUCCUUUAUUUCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUACAGGACUCGCGUACCUUCAUUGGCACCUUCAAAGCCUU --........................(((..............(((((.((((((((....)))))))))))))((((((.(((((....)).)))....)))))).......))).. ( -33.40, z-score = -2.85, R) >droAna3.scaffold_12916 14267722 106 - 16180835 ------------UAUUUCAGACUAUCGGAAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUUCUCCAGGACUCUCGCACGUUCAUUGGGACGUUCAAGGCCUU ------------..............((...((((........(((((.((((((((....)))))))))))))(((((...((((........))))...)))))))))....)).. ( -32.00, z-score = -2.66, R) >droPer1.super_1 9293949 114 + 10282868 ----AUUAUUUUACUUACAGACUAUUGGGAAGAAUAACAAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUCUGCAAGACUCACGUACAUUCAUCGGAACUUUCAAAGCAUU ----.(((((((.((((........)))).)))))))......(((((.(((((((......))))))))))))((((((...((...........))..))))))............ ( -22.80, z-score = -1.38, R) >dp4.chr4_group2 192496 114 + 1235136 ----AUUAUUUUACUUACAGACUAUUGGGAAGAAUAACAAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUCUGCAAGACUCACGUACAUUCAUCGGAACUUUCAAAGCAUU ----.(((((((.((((........)))).)))))))......(((((.(((((((......))))))))))))((((((...((...........))..))))))............ ( -22.80, z-score = -1.38, R) >droWil1.scaffold_180708 4863903 114 - 12563649 ----UUUUUCUUUCUUUCAGACUAUUGGAAAGAAUAAUAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUAUUACAAGAUUCUCGUACGUUCAUUGGAACUUUUAAGGCUUU ----...((((((((...........))))))))...(((.(((((((.((((((((....))))))))))))))).)))..............((((....))))............ ( -29.60, z-score = -3.15, R) >droVir3.scaffold_12963 19183973 116 + 20206255 -UAUGCU-CCCAUUUUUCAGACCAUAGGCAAGAAUAACAAAAUGAUUCAGCACCUGAACUAUCGGGUGCGAAUUGUGCUGCAGGAUUCGAGGACGUUUAUUGGAACUUUCAAGGCAUU -.(((((-.......................((((..((..(..((((.((((((((....))))))))))))..)..))....))))..(((.((((....)))).)))..))))). ( -28.10, z-score = -0.61, R) >droMoj3.scaffold_6500 4600120 117 + 32352404 UUUUUUU-UUUUUUUUUUAGACUAUUGGCAAGAAUAAUAAAAUGAUUCAGCACCUGAACUAUCGGGUGCGGAUUGUUCUGCAGGAUUCAAGAACUUUUAUUGGAACAUUUAAGGCAUU .......-...............................(((((.((((((((((((....)))))))).((..(((((..........)))))..))..)))).)))))........ ( -23.00, z-score = -1.04, R) >droGri2.scaffold_15126 2426145 118 - 8399593 UUAUAUUAUUCCUUUCGCAGACUAUUGGCAAGAAUAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGAUUCGAGGACAUUCAUCGGCACAUUCAAAGCAUU ...........................((..((((........(((((.((((((((....)))))))))))))((((((..((((........))))..))))))))))...))... ( -31.60, z-score = -2.30, R) >anoGam1.chr2L 32754160 117 + 48795086 -ACAUUCCCUCUCCUUACAGACGAUCGGCAAAAAUAACAAAAUGAUCCAGCUCCUGAACUACAGGGUGCGGAUCGUUUUGCAGGACUCGCGUACCUUCAUCGGCACGUUCAAAGCGUU -.....................((.((((........(((((((((((.(((((((.....))))).))))))))))))).(((((....)).)))......)).)).))........ ( -30.20, z-score = -1.24, R) >apiMel3.GroupUn 18893712 117 + 399230636 -UAGAAACAUUUUUUUUCAGACGAUUGGUAAAAAUAAUAAAAUGCUACAACAUAUUAAUUAUAGAGUCAGAAUUAUUUUGCAAGAUUCAAGGACAUUCAUAGGCACAUUUAAAGCCUU -..(((.((((((.......((.....)).........)))))).....................(((.(((((.........)))))...))).)))..((((.........)))). ( -10.99, z-score = 0.83, R) >triCas2.ChLG5 9637801 116 - 18847211 --CACCGUAUUUCCUUUCAGACGAUGGGCAAAAACAACAAAAUGUUACAACACAUCAACUACCGAGUCCGGAUAACCCUCCAAGACUCCCGGACCUUCAUCGGCACAUUCAAGGCCUU --..(((...............((((......((((......))))......)))).......(((((.(((......)))..))))).))).........(((.(......)))).. ( -21.30, z-score = -0.62, R) >consensus __UGAAUUCCUUUCUUUCAGACGAUCGGCAAGAAUAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCGUACCUUCAUCGGAACCUUCAAAGCAUU ...........................................(((((.(((((((......))))))))))))((((((...((...........))..))))))............ (-15.64 = -16.07 + 0.43)
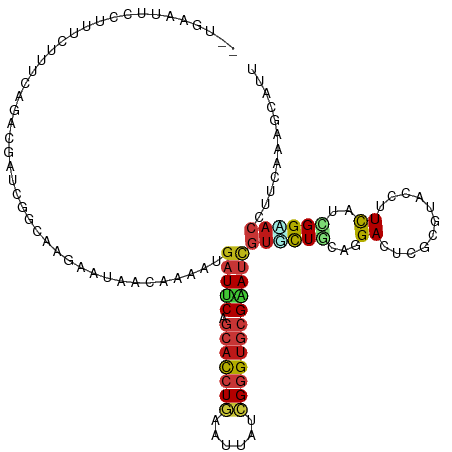
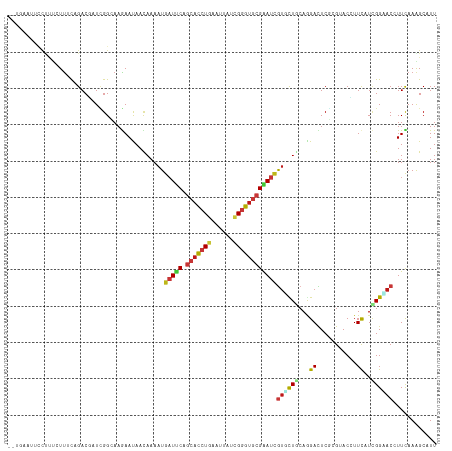
| Location | 10,426,066 – 10,426,162 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.93 |
| Shannon entropy | 0.63768 |
| G+C content | 0.36673 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -11.62 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10426066 96 + 23011544 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUG-AAAUAAAGGAAUUCAAAGGAUUUAAUUGUGAAAUA-------------------- .((((((..(((((.((....)).)))))((((((.(((((..((((((.....((....)-).....)))))).))))))))))))))))).....-------------------- ( -25.10, z-score = -2.00, R) >droSim1.chr2L 10217855 96 + 22036055 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUG-CAAAAAAGGAGUUCACAGGAUUAAAUUGUGAAAUA-------------------- .(((((((((((((.((....)).))))).)))))......(((.(((((..((.(..((.-......))..).)).))))).)))...))).....-------------------- ( -25.10, z-score = -1.49, R) >droSec1.super_3 5853966 96 + 7220098 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUG-CAACAAAGGAGUUCAAAGGAUUUAAUUGUGAAAUA-------------------- .(((((((.(((((.((....)).)))))(((((..(((((((((...((.....))...)-))))))))..)))))........))))))).....-------------------- ( -28.10, z-score = -2.43, R) >droYak2.chr2L 13106592 96 + 22324452 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUUGUCUA-AAACAAAGAAAUACAAAGGAUUUAAUUGUGAAAUA-------------------- .((((((..(((((.((....)).)))))((((((.((((((..(((((............-.....)))))..)))))))))))))))))).....-------------------- ( -24.93, z-score = -2.88, R) >droEre2.scaffold_4929 11035367 96 + 26641161 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUG-AAAUAAAGGAGUUCAAGGGAUUUAAUCGUGAAAUA-------------------- .(((((((.(((((.((....)).)))))(((((..(((((((..((..((.....))..)-))))))))..)))))........))))))).....-------------------- ( -24.90, z-score = -1.26, R) >droAna3.scaffold_12916 14267762 78 + 16180835 GAACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUUCCGAUAGUCUG-AAAUAAAUGGUGAAAUA-------------------------------------- ....((((((((((.((....)).))))).)))))((((..(..((.((((.(.....).)-)))..))..)..)))).-------------------------------------- ( -21.50, z-score = -2.94, R) >droPer1.super_1 9293989 90 - 10282868 GAACGAUUCGCACCCGGUAGUUCAGAUGCUGAAUCAUUUUGUUAUUCUUCCCAAUAGUCUGUAAGUAAAAUAAUAUUAGGUUAACAAAUG--------------------------- ....((((((((.(.((....)).).))).)))))..(((((((.....((.((((.................)))).)).)))))))..--------------------------- ( -13.23, z-score = -0.25, R) >dp4.chr4_group2 192536 90 - 1235136 GAACGAUUCGCACCCGGUAGUUCAGAUGCUGAAUCAUUUUGUUAUUCUUCCCAAUAGUCUGUAAGUAAAAUAAUAUUAAGUUAACAAAUG--------------------------- ....((((((((.(.((....)).).))).)))))..(((((((..(((...((((.................))))))).)))))))..--------------------------- ( -11.13, z-score = 0.02, R) >droWil1.scaffold_180708 4863943 94 + 12563649 AUACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUAUUAUUCUUUCCAAUAGUCUG-AAAGAAAGAAAA--AAGAAAAUUAAUCACAAAAUA-------------------- ....((((((((((.((....)).))))).))))).((((.((.(((((((.........)-)))))).)).))--))...................-------------------- ( -21.10, z-score = -3.97, R) >droMoj3.scaffold_6500 4600160 100 - 32352404 GAACAAUCCGCACCCGAUAGUUCAGGUGCUGAAUCAUUUUAUUAUUCUUGCCAAUAGUCUAAAAAAAAAAAAAAA-AAUAGAAACAUUUCCA--UUAAAAACA-------------- ......((.(((((.((....)).))))).))...........................................-................--.........-------------- ( -11.10, z-score = -1.07, R) >droGri2.scaffold_15126 2426185 117 + 8399593 GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUAUUCUUGCCAAUAGUCUGCGAAAGGAAUAAUAUAAUGAGAGAAAUUCAAAAUAGAAAACAUUUAACCAUAUUUU ....((((((((((.((....)).))))).)))))((((((...(((((..(((((...(.(....).)....)))..)).)))))...))))))...................... ( -23.90, z-score = -1.93, R) >anoGam1.chr2L 32754200 102 - 48795086 AAACGAUCCGCACCCUGUAGUUCAGGAGCUGGAUCAUUUUGUUAUUUUUGCCGAUCGUCUGUAAGGAGAGGGAAUGUUGGAGGAUAAACUCAUUGCGAAAAG--------------- ....(((((((..((((.....)))).)).))))).........(((((((((((..(((.(......).)))..))))(((......)))...))))))).--------------- ( -26.80, z-score = -0.71, R) >triCas2.ChLG5 9637841 82 + 18847211 GGGUUAUCCGGACUCGGUAGUUGAUGUGUUGUAACAUUUUGUUGUUUUUGCCCAUCGUCUGAAAGGAAAUACGGUGGAAAUG----------------------------------- (((((.....))))).......(((((......))))).....(((((..((.....(((....))).....))..))))).----------------------------------- ( -18.00, z-score = -0.15, R) >consensus GCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUAGUCUG_AAAUAAAGGAAUACAAAGGAAUUAAUUGUGAAAUA____________________ ....((((((((((.((....)).))))).))))).................................................................................. (-11.62 = -12.15 + 0.52)

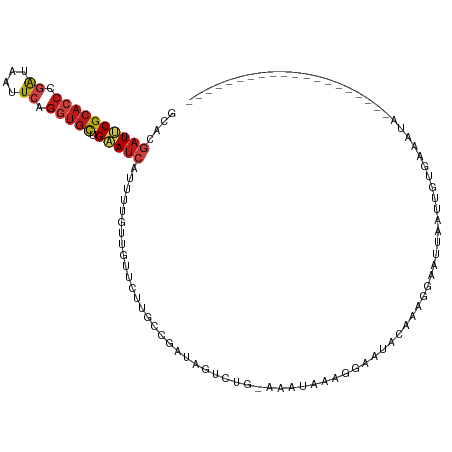
| Location | 10,426,066 – 10,426,162 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.93 |
| Shannon entropy | 0.63768 |
| G+C content | 0.36673 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10426066 96 - 23011544 --------------------UAUUUCACAAUUAAAUCCUUUGAAUUCCUUUAUUU-CAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC --------------------.....(((......(((.((((((.........))-)))).)))..................(((((.((((((((....)))))))))))))))). ( -25.20, z-score = -3.17, R) >droSim1.chr2L 10217855 96 - 22036055 --------------------UAUUUCACAAUUUAAUCCUGUGAACUCCUUUUUUG-CAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC --------------------...((((((.........)))))).....((((((-(.(.....).))))))).......(((((((.((((((((....))))))))))))))).. ( -29.80, z-score = -3.94, R) >droSec1.super_3 5853966 96 - 7220098 --------------------UAUUUCACAAUUAAAUCCUUUGAACUCCUUUGUUG-CAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC --------------------.....(((......(((.(((((((......))).-)))).)))..................(((((.((((((((....)))))))))))))))). ( -25.50, z-score = -2.53, R) >droYak2.chr2L 13106592 96 - 22324452 --------------------UAUUUCACAAUUAAAUCCUUUGUAUUUCUUUGUUU-UAGACAAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC --------------------.....(((..........(((((..(((((.(((.-.........))))))))..)))))..(((((.((((((((....)))))))))))))))). ( -25.50, z-score = -3.46, R) >droEre2.scaffold_4929 11035367 96 - 26641161 --------------------UAUUUCACGAUUAAAUCCCUUGAACUCCUUUAUUU-CAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC --------------------.......(((((........((((....))))...-.....)))))..............(((((((.((((((((....))))))))))))))).. ( -23.39, z-score = -2.23, R) >droAna3.scaffold_12916 14267762 78 - 16180835 --------------------------------------UAUUUCACCAUUUAUUU-CAGACUAUCGGAAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUUC --------------------------------------..............(((-(.(.....).)))).........((((((((.((((((((....)))))))))))))))). ( -21.50, z-score = -3.89, R) >droPer1.super_1 9293989 90 + 10282868 ---------------------------CAUUUGUUAACCUAAUAUUAUUUUACUUACAGACUAUUGGGAAGAAUAACAAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUC ---------------------------..(((((((.(((((((.................))))))).....)))))))(((((((.(((((((......)))))))))))))).. ( -23.23, z-score = -3.82, R) >dp4.chr4_group2 192536 90 + 1235136 ---------------------------CAUUUGUUAACUUAAUAUUAUUUUACUUACAGACUAUUGGGAAGAAUAACAAAAUGAUUCAGCAUCUGAACUACCGGGUGCGAAUCGUUC ---------------------------..(((((((.(((..((.((.(((......))).)).))..)))..)))))))(((((((.(((((((......)))))))))))))).. ( -20.50, z-score = -3.01, R) >droWil1.scaffold_180708 4863943 94 - 12563649 --------------------UAUUUUGUGAUUAAUUUUCUU--UUUUCUUUCUUU-CAGACUAUUGGAAAGAAUAAUAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUAU --------------------.....................--..((((((((..-.........)))))))).......(((((((.((((((((....))))))))))))))).. ( -26.50, z-score = -4.34, R) >droMoj3.scaffold_6500 4600160 100 + 32352404 --------------UGUUUUUAA--UGGAAAUGUUUCUAUU-UUUUUUUUUUUUUUUAGACUAUUGGCAAGAAUAAUAAAAUGAUUCAGCACCUGAACUAUCGGGUGCGGAUUGUUC --------------(((((((((--(((((....)))))))-..................(....)..)))))))....((..((((.((((((((....))))))))))))..)). ( -20.80, z-score = -1.85, R) >droGri2.scaffold_15126 2426185 117 - 8399593 AAAAUAUGGUUAAAUGUUUUCUAUUUUGAAUUUCUCUCAUUAUAUUAUUCCUUUCGCAGACUAUUGGCAAGAAUAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC (((((((......))))))).(((..(((.......)))..)))((((((...(((........)))...))))))....(((((((.((((((((....))))))))))))))).. ( -24.70, z-score = -1.67, R) >anoGam1.chr2L 32754200 102 + 48795086 ---------------CUUUUCGCAAUGAGUUUAUCCUCCAACAUUCCCUCUCCUUACAGACGAUCGGCAAAAAUAACAAAAUGAUCCAGCUCCUGAACUACAGGGUGCGGAUCGUUU ---------------......((...(((......)))........(.(((......))).)....))..........(((((((((.(((((((.....))))).))))))))))) ( -21.30, z-score = -1.08, R) >triCas2.ChLG5 9637841 82 - 18847211 -----------------------------------CAUUUCCACCGUAUUUCCUUUCAGACGAUGGGCAAAAACAACAAAAUGUUACAACACAUCAACUACCGAGUCCGGAUAACCC -----------------------------------........(((.((((......((..((((......((((......))))......))))..))...)))).)))....... ( -9.10, z-score = 0.58, R) >consensus ____________________UAUUUCACAAUUAAAUCCUUUGAAUUCCUUUAUUU_CAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGC ................................................................................(((((((.(((((((......)))))))))))))).. (-16.08 = -16.40 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:06 2011