| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,321,551 – 6,321,653 |
| Length | 102 |
| Max. P | 0.790154 |

| Location | 6,321,551 – 6,321,653 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Shannon entropy | 0.45681 |
| G+C content | 0.42245 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.24 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6321551 102 - 22422827 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGG-------UUUCUCAGGAUUUUUCCGGCUCUU- .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((((((((.-------...))).))))...)))))))...- ( -31.20, z-score = -2.08, R) >droEre2.scaffold_4690 15276647 103 - 18748788 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAACCGGGGUCCGGGG-------UUUUUCAGGGGUUUUCAGGGUUUUU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))(((((((..((..((-------....))..))..)))...))))... ( -20.60, z-score = 0.57, R) >droYak2.chrX 2306766 109 - 21770863 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGAGUUCUUUUUUUUUCCAGGCUUUUCCGGCUUCU- .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((((.((((..........)))).))))...))))))...- ( -31.90, z-score = -2.07, R) >droSec1.super_4 5721814 101 + 6179234 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGG-------UUUUUCAGGG-UUUUCCGGCUCUU- .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((..((-------....))..))-..))))))))...- ( -29.30, z-score = -1.68, R) >droSim1.chrX 4965501 101 - 17042790 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGG-------UUUUUCAGGG-UUUUCCGGCUCUU- .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((..((-------....))..))-..))))))))...- ( -29.30, z-score = -1.68, R) >droWil1.scaffold_181150 55284 84 + 4952429 -UCGUUGGCUGAGUGAAUGGAUUAAAAGUCCAUCAAAAUGUACAUAAAAGUCAUUGACUUAACAGGCAACGUCUCUG--------GAUAUCAG----------------- -.(((((.(((..(((.(((((.....))))))))............(((((...)))))..))).)))))......--------........----------------- ( -20.50, z-score = -2.17, R) >droMoj3.scaffold_6473 14536073 96 + 16943266 ------GCGCGUAUGUGUGGAUUAAAAGUCCAUCAAAAUGUACAUAAAAGUCAUUGACUUAACAGGCAGCUGCAAGC-----CUGGUUACCAAG--UUAUGCGACACCU- ------.((((((....(((.....(((((......((((.((......)))))))))))..(((((........))-----)))....)))..--.))))))......- ( -24.10, z-score = -0.98, R) >droAna3.scaffold_13335 1292199 87 + 3335858 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCUGGA--------UACCGGCGGG--------------- .......(((((((((.(((((.....)))))))..((((.((......)))))).))))))).(((((..(....)--------..)))))...--------------- ( -25.00, z-score = -1.95, R) >consensus UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGG_______UUUUUCAGGG_UUUUCCGGCUCUU_ ....(((((((..((.((((((.....))))))))............(((((...)))))..)))))))......................................... (-15.28 = -15.24 + -0.05)

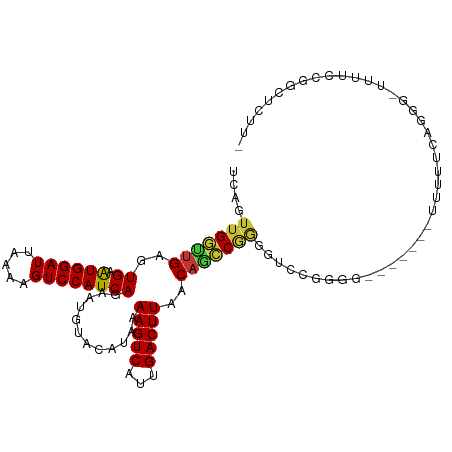
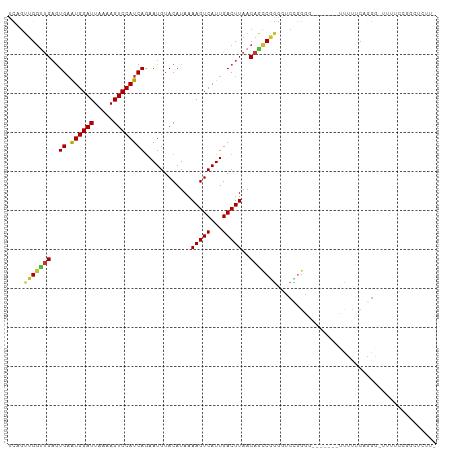
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:37 2011