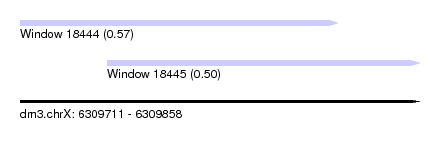
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,309,711 – 6,309,858 |
| Length | 147 |
| Max. P | 0.570373 |

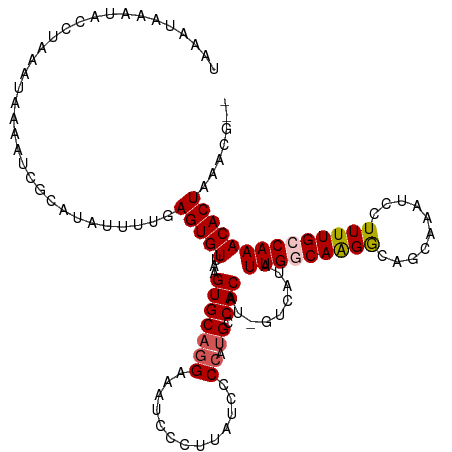
| Location | 6,309,711 – 6,309,828 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.36671 |
| G+C content | 0.38372 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6309711 117 + 22422827 UAAAUAAAUACCAAAAUAGAAUCGCAUAUUUUGAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAUCGUCAUAUUGGCAAGGCAACAAAUCCUUUUGCCAAACACUAAACG-- ......((((((((((((........))))))).))))).((((.(((.....)))........(((((...........))))).(((((.........)))))...)))).....-- ( -22.70, z-score = -2.18, R) >droSim1.chrX 4951429 111 + 17042790 UAAAUAAAUGCCUAAAUAAAAUCGCAUAUUUUGAGUGUUAUGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU------AUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG-- ........(((((..........((((((..........))))))(((.....)))........(((((...------..))))))))))(((((....))))).............-- ( -23.40, z-score = -1.83, R) >droSec1.super_4 5709400 111 - 6179234 UAAAUAAAUAGCUAAAUGAAAUCGCAUAUUUUGAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU------AUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG-- .....(((((((...........)).)))))..((((((...((((((.....)))........(((((...------..)))))..)))(((((....)))))..)))))).....-- ( -20.70, z-score = -0.83, R) >droYak2.chrX 2296615 109 + 21770863 ---------GAAUAAAUAGAAUUCUAUAUUUUGAGUGUUGAGUGCAGGAAAUUUCUCAUUCCC-UGCCACAUUGUCAUAUUGACAGGACAGCAAAUCCUUUUGCCAAACACUAAACGCU ---------......((((....))))......((((((..(((((((.(((.....))).))-)).))).(((((.....)))))....(((((....)))))..))))))....... ( -23.70, z-score = -1.73, R) >droEre2.scaffold_4690 15263919 117 + 18748788 CUCGUGAACACUCAAAUAAAAACCUCUAUUUUGAGUGUCAAGUGCAGGAAUUCCCUAUCCCCAAAGGCACAUCGUCAUAUUGCCAAGACAGCAAAUCCUUUUGCCAAACACUAAACG-- ....(((.((((((((.............)))))))))))((((.(((.....))).........((((...........))))......(((((....)))))....)))).....-- ( -23.12, z-score = -2.50, R) >consensus UAAAUAAAUACCUAAAUAAAAUCGCAUAUUUUGAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU_GUCAUAUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG__ .................................(((((...(((((((.............)).)).))).........(((((((((..........))))))))))))))....... (-12.44 = -12.84 + 0.40)

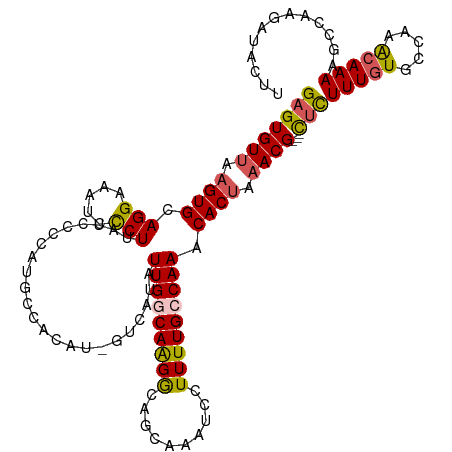
| Location | 6,309,743 – 6,309,858 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Shannon entropy | 0.31073 |
| G+C content | 0.43038 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 6309743 115 + 22422827 GAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAUCGUCAUAUUGGCAAGGCAACAAAUCCUUUUGCCAAACACUAAACG---CUCUUUGUGCCAAACAAAAGCCA-GAUACUU (((((((.((((.(((.....)))........(((((...........))))).(((((.........)))))...)))).))))---)))(((((.....))))).....-....... ( -27.30, z-score = -2.29, R) >droSim1.chrX 4951461 110 + 17042790 GAGUGUUAUGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU------AUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG---CUCUUUGUGCCAAACAAAAGCCAAGAUACUU (((((((..(((.(((.....))).....((.(((((...------..))))).))..(((((....)))))....)))..))))---)))(((((.....)))))............. ( -26.40, z-score = -1.59, R) >droSec1.super_4 5709432 110 - 6179234 GAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU------AUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG---CUCUUUGUGCCAAACAAAGGCCAAGAUACUU (((((((.((((.(((.....))).....((.(((((...------..))))).))..(((((....)))))....)))).))))---)))((((.(((.......)))))))...... ( -32.50, z-score = -3.07, R) >droYak2.chrX 2296638 111 + 21770863 GAGUGUUGAGUGCAGGAAAUUUCUCAUUCCC-UGCCACAUUGUCAUAUUGACAGGACAGCAAAUCCUUUUGCCAAACACUAAACGCUUUUUUUUUUGUCAAACAAAAGCUAA------- .((((((..(((((((.(((.....))).))-)).))).(((((.....)))))....(((((....)))))..))))))....((((((.(((.....))).))))))...------- ( -26.50, z-score = -2.06, R) >droEre2.scaffold_4690 15263951 116 + 18748788 GAGUGUCAAGUGCAGGAAUUCCCUAUCCCCAAAGGCACAUCGUCAUAUUGCCAAGACAGCAAAUCCUUUUGCCAAACACUAAACG---UGCUUUGUGGCAAGCAAAACCCCAGAUACUU (((((((..((((.(((........)))......)))).........((((((.....(((((....)))))(((((((.....)---)).))))))))))...........))))))) ( -29.00, z-score = -2.28, R) >consensus GAGUGUUAAGUGCAGGAAAUCCCUUAUCCCCAUGCCACAU_GUCAUAUUGGCAAGGCAGCAAAUCCUUUUGCCAAACACUAAACG___CUCUUUGUGCCAAACAAAAGCCAAGAUACUU .(((((...(((((((.............)).)).))).........(((((((((..........))))))))))))))...........(((((.....)))))............. (-13.80 = -14.24 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:35 2011