| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,307,881 – 6,307,986 |
| Length | 105 |
| Max. P | 0.909198 |

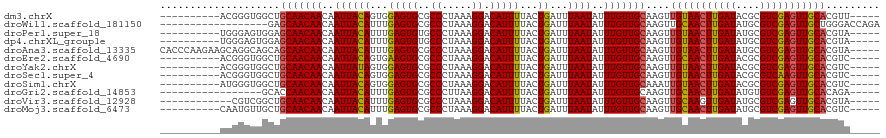
| Location | 6,307,881 – 6,307,986 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.38 |
| Shannon entropy | 0.26174 |
| G+C content | 0.40760 |
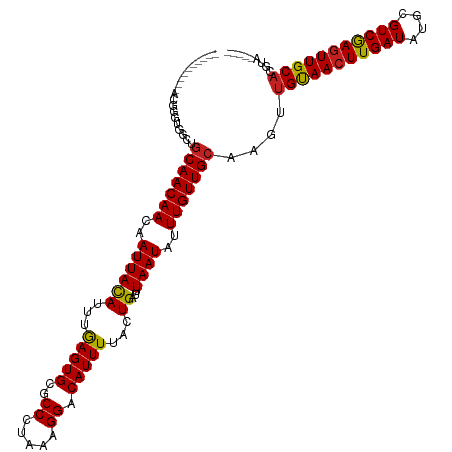
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6307881 105 + 22422827 ----------ACGGGUGGCUGCAACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUU----- ----------(((...(((((((((((..((((((((((((((..((.....)).))))))))))...))))..))))))).))))((((((((((....)))))))))).))).----- ( -35.30, z-score = -2.96, R) >droWil1.scaffold_181150 41310 102 - 4952429 ------------------GAGCAACAACAAUUACAUUUGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGCAACUUGAUAUGCGUCGAGUUGCUGGGACCAGA ------------------..(((((((..((((.(((.(((((..((.....)).)))))....))).))))..))))))).....((((((((((....)))))))))).......... ( -26.70, z-score = -1.75, R) >droPer1.super_18 1734019 105 + 1952607 ----------UGGGAGUGGAGCAACAACAAUUACAUUUGAGUGUGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUAUGCGUCGAGUUGCACGUA----- ----------..........(((((((..((((.(((.((((((.((.....))))))))....))).))))..)))))))..((.((((((((((....))))))))))))...----- ( -28.30, z-score = -1.77, R) >dp4.chrXL_group1e 2843962 105 + 12523060 ----------UGGGAGUGGAGCAACAACAAUUACAUUUGAGUGUGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUAUGCGUCGAGUUGCACGUA----- ----------..........(((((((..((((.(((.((((((.((.....))))))))....))).))))..)))))))..((.((((((((((....))))))))))))...----- ( -28.30, z-score = -1.77, R) >droAna3.scaffold_13335 1276167 115 - 3335858 CACCCAAGAAGCAGGCAGCAGCAACAACAAUUACAUUUGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUAUGCGUCGAGUUGCACGUA----- ..........((.....)).(((((((..((((.(((.(((((..((.....)).)))))....))).))))..)))))))..((.((((((((((....))))))))))))...----- ( -27.10, z-score = -0.61, R) >droEre2.scaffold_4690 15262171 105 + 18748788 ----------ACGGGUGGCUGCAACAACAAUUACAGUGAAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGCAACUUGAUACGCGUCGAGUUGCACGUC----- ----------(((...(((((((((((..((((((((((((((..((.....)).))))))))))...))))..))))))).))))((((((((((....)))))))))).))).----- ( -37.70, z-score = -3.71, R) >droYak2.chrX 2294793 105 + 21770863 ----------ACGGGUGGCUGCAACAACAAUUAUAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUC----- ----------(((...(((((((((((..((((((((((((((..((.....)).))))))))))...))))..))))))).))))((((((((((....)))))))))).))).----- ( -33.70, z-score = -2.61, R) >droSec1.super_4 5707511 105 - 6179234 ----------ACGGGUGGCUGCAACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCAAGUUGCACGUC----- ----------(((...(((((((((((..((((((((((((((..((.....)).))))))))))...))))..))))))).))))((((((((((....)))))))))).))).----- ( -35.60, z-score = -3.14, R) >droSim1.chrX 4949648 105 + 17042790 ----------AUGGGUGGCUGCAACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAAUUGUAACUUGAUACGCGUCGAGUUGCACGUC----- ----------......(((((((((((..((((((((((((((..((.....)).))))))))))...))))..))))))))...(((((((((((....))))))))))).)))----- ( -34.10, z-score = -3.00, R) >droGri2.scaffold_14853 588176 98 - 10151454 -----------------GCACCAACAACAAUUACAUUUGAGUGCGCCCUUAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGCAACUUGAUAUGUGUCGAGUUGCACAGA----- -----------------....((((((..((((.(((.(((((..((.....)).)))))....))).))))..))))))...((.((((((((((....))))))))))))...----- ( -23.50, z-score = -1.51, R) >droVir3.scaffold_12928 7601855 103 + 7717345 ------------CGUCGGCUGCAACAACAAUUACAUUUGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGCAAGUUGAUAUGCGUCGAGUUGCACGUA----- ------------(((.(((((((((((..((((.(((.(((((..((.....)).)))))....))).))))..))))))).))))((((.(((((....))))).)))))))..----- ( -25.80, z-score = -0.44, R) >droMoj3.scaffold_6473 14519780 105 - 16943266 ----------CAAUGUUGCUGCAACAACAAUUACAUUUGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGCAACUUGAUAUGCGUCGAGUUGCACGUC----- ----------.......((((((((((..((((.(((.(((((..((.....)).)))))....))).))))..))))))).)))(((((((((((....)))))))))))....----- ( -28.60, z-score = -1.73, R) >consensus __________ACGGGUGGCUGCAACAACAAUUACAUUUGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUAUGCGUCGAGUUGCACGUA_____ ....................(((((((..((((((...(((((..((.....)).)))))...))...))))..)))))))....(((((((((((....)))))))))))......... (-23.15 = -22.93 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:32 2011