
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,283,078 – 6,283,156 |
| Length | 78 |
| Max. P | 0.606802 |

| Location | 6,283,078 – 6,283,156 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 71.62 |
| Shannon entropy | 0.53829 |
| G+C content | 0.57638 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -13.08 |
| Energy contribution | -15.20 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
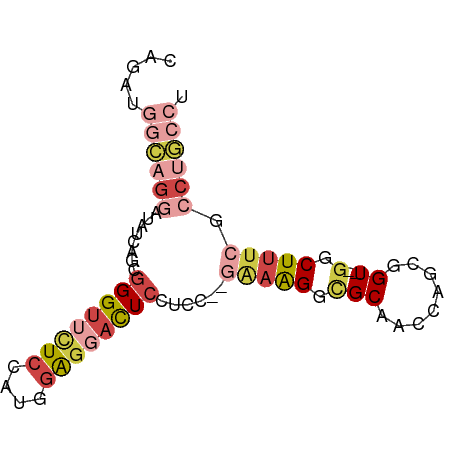
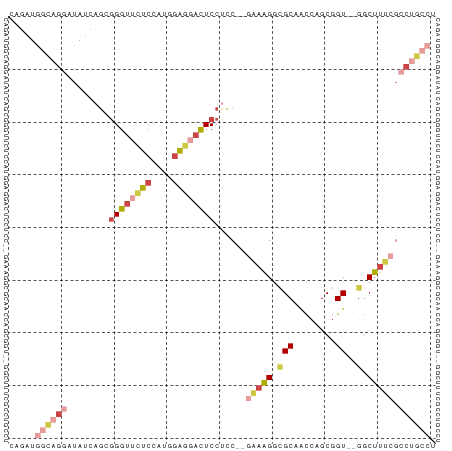
>dm3.chrX 6283078 78 + 22422827 CAGAUGGCAGGAUAUCAGCGGGUUCUCCAUGGAGGACUCCUCCC-AAAAGGCGCAACCGGCGGU--GGCUUUCGCCUGCCU .....((((((.....(((((((((((....)))))))).....-......(((.....)))..--.)))....)))))). ( -30.10, z-score = -0.73, R) >droSim1.chrX_random 2042575 77 + 5698898 CAGAUGGCAGGAUAUCAGUGGGUUCUCCAUGGAGCACUCCUCC--GAAAGGCGCAACCAGCGGU--GGCUUUCGCCUGCCU .....((((((.....((.((((.(((....))).)))))).(--(((((.(((........))--).)))))))))))). ( -31.30, z-score = -1.62, R) >droSec1.super_4 5689156 77 - 6179234 CAGAUGGCAGGAUAUCAGUGGGUUCUCCAUGGAGCACUCCUCC--GAAAGGCGCAACCAGCGGU--GGCUUUCGCCUGCCU .....((((((.....((.((((.(((....))).)))))).(--(((((.(((........))--).)))))))))))). ( -31.30, z-score = -1.62, R) >droYak2.chrX 2275897 77 + 21770863 CAGAUGGCAGGAUAUCAGGGGGUUUUCCAUGGAGGACUCCUCC--GCAAGGAGCAACCAGCGGU--GGCUUUCGCCUGCCU .....((((((.......(((((((((....)))))))))(((--....)))((.(((...)))--.)).....)))))). ( -33.30, z-score = -1.96, R) >droEre2.scaffold_4690 15243863 70 + 18748788 CAGAUGGCUGGGUAUCAGGGGGUUUUCCAUGGAGGACUCCAUGGAGAAAGGCGCGGCCAGCGGU--GGCUUU--------- ....((((((.((.((..(((((((((....)))))))))...)).....)).))))))((...--.))...--------- ( -28.20, z-score = -2.42, R) >droAna3.scaffold_13335 1237640 71 - 3335858 ----UAUAUGUAUAUAUACAGGAUAUCUCUUAGCGAUUCCU----GGUGGCUGCCGUUUGCCGUCUGCCUUUCGCCUUU-- ----....((((....))))............((((.....----(..(((........)))..)......))))....-- ( -13.20, z-score = 0.43, R) >consensus CAGAUGGCAGGAUAUCAGCGGGUUCUCCAUGGAGGACUCCUCC__GAAAGGCGCAACCAGCGGU__GGCUUUCGCCUGCCU .....((((((........((((((((....))))))))......(((((.(...((.....))..).))))).)))))). (-13.08 = -15.20 + 2.12)

| Location | 6,283,078 – 6,283,156 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 71.62 |
| Shannon entropy | 0.53829 |
| G+C content | 0.57638 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -9.33 |
| Energy contribution | -11.17 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
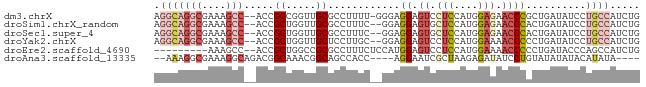
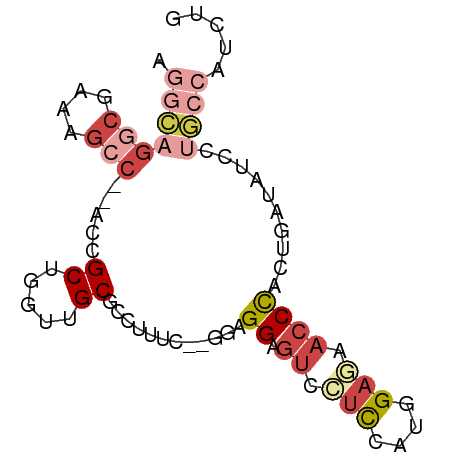
>dm3.chrX 6283078 78 - 22422827 AGGCAGGCGAAAGCC--ACCGCCGGUUGCGCCUUUU-GGGAGGAGUCCUCCAUGGAGAACCCGCUGAUAUCCUGCCAUCUG .(((((((....)))--...((.((((.(.((....-.(((((...)))))..)).))))).))........))))..... ( -31.10, z-score = -1.62, R) >droSim1.chrX_random 2042575 77 - 5698898 AGGCAGGCGAAAGCC--ACCGCUGGUUGCGCCUUUC--GGAGGAGUGCUCCAUGGAGAACCCACUGAUAUCCUGCCAUCUG .((((((((..((((--(....))))).))))..((--((.((.((.(((....))).)))).)))).....))))..... ( -29.10, z-score = -1.53, R) >droSec1.super_4 5689156 77 + 6179234 AGGCAGGCGAAAGCC--ACCGCUGGUUGCGCCUUUC--GGAGGAGUGCUCCAUGGAGAACCCACUGAUAUCCUGCCAUCUG .((((((((..((((--(....))))).))))..((--((.((.((.(((....))).)))).)))).....))))..... ( -29.10, z-score = -1.53, R) >droYak2.chrX 2275897 77 - 21770863 AGGCAGGCGAAAGCC--ACCGCUGGUUGCUCCUUGC--GGAGGAGUCCUCCAUGGAAAACCCCCUGAUAUCCUGCCAUCUG .(((((((....)))--.(((.(((..(((((((..--.)))))))...)))))).................))))..... ( -29.20, z-score = -2.11, R) >droEre2.scaffold_4690 15243863 70 - 18748788 ---------AAAGCC--ACCGCUGGCCGCGCCUUUCUCCAUGGAGUCCUCCAUGGAAAACCCCCUGAUACCCAGCCAUCUG ---------......--...(((((...((......((((((((....))))))))........))....)))))...... ( -18.84, z-score = -2.41, R) >droAna3.scaffold_13335 1237640 71 + 3335858 --AAAGGCGAAAGGCAGACGGCAAACGGCAGCCACC----AGGAAUCGCUAAGAGAUAUCCUGUAUAUAUACAUAUA---- --...(.(....).)....(((........)))..(----(((((((.......))).)))))..............---- ( -15.90, z-score = -1.40, R) >consensus AGGCAGGCGAAAGCC__ACCGCUGGUUGCGCCUUUC__GGAGGAGUCCUCCAUGGAGAACCCACUGAUAUCCUGCCAUCUG .((((((.............((.....))............((.((.(((....))).))))........))))))..... ( -9.33 = -11.17 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:29 2011