
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,420,849 – 10,420,921 |
| Length | 72 |
| Max. P | 0.729904 |

| Location | 10,420,849 – 10,420,921 |
|---|---|
| Length | 72 |
| Sequences | 11 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 50.71 |
| Shannon entropy | 1.03994 |
| G+C content | 0.47361 |
| Mean single sequence MFE | -16.99 |
| Consensus MFE | -5.62 |
| Energy contribution | -6.10 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.88 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.729904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
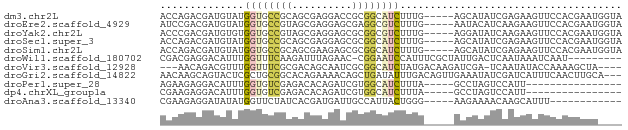
>dm3.chr2L 10420849 72 - 23011544 ACCAGACGAUGUAUGGUGCCGCAGCGAGGACCGCGGCAUCUUUG-----AGCAUAUCGAGAAGUUCCACGAAUGGUA ((((..((......((((((((.(......).))))))))((((-----(.....)))))........))..)))). ( -21.90, z-score = -0.96, R) >droEre2.scaffold_4929 11030366 72 - 26641161 AUCCGACGAUGUAUGGUGCCGUAGCGAGGAGCGAGGCGUCUUUG-----AAUACAUCAAGAAGUUCCACGAAUGGUA .......((((((((..(((...((.....))..)))..)....-----.)))))))........(((....))).. ( -17.30, z-score = 0.20, R) >droYak2.chr2L 13101519 72 - 22324452 ACCCGACGAUGUGUGGUGCCGUAGCGAGGAGCGCGGCGUCUUUG-----AGGAUAUCAAGAAGUUCCACGAAUGGUA .....((.((.(((((.(((((.((.....))))))).((((.(-----(.....))))))....))))).)).)). ( -21.30, z-score = -0.28, R) >droSec1.super_3 5848715 72 - 7220098 ACCAGACGAUGUAUGGUGCCGCAGCGAGGAGCGCGGCAUCUUUG-----AGCAUAUCGAGAAGUUCCACGAAUGGUA ((((..((......((((((((.((.....))))))))))((((-----(.....)))))........))..)))). ( -25.20, z-score = -1.95, R) >droSim1.chr2L 10212612 72 - 22036055 ACCAGACGAUGUAUGGUGCCGCAGCGAAGAGCGCGGCAUCUUUG-----AGCAUAUCGAGAAGUUCCACGAAUGGUA ((((..((......((((((((.((.....))))))))))((((-----(.....)))))........))..)))). ( -25.20, z-score = -2.27, R) >droWil1.scaffold_180702 3707749 67 + 4511350 CGACGAGGACAUUUGGUUUCAAGAUUUAGAAC-CGGAAUCCAUUUCGCUAUUGACUCAAUAAAUCAAU--------- (((...(((..((((((((.........))))-)))).)))...))).(((((...))))).......--------- ( -9.40, z-score = 0.29, R) >droVir3.scaffold_12928 7397811 69 + 7717345 ---AACAGACGUUUGGUUUCGCGACAGCAAUCGCGGCAUCUAUGACAAGAUCGA-UCAAUAUACCAAAAGCUA---- ---...((...((((((..(((((......)))))(.((((......)))))..-.......))))))..)).---- ( -14.10, z-score = -0.90, R) >droGri2.scaffold_14822 209400 74 - 1097346 AACAAGCAGUACUCGCUGCGGCACAGAAAACAGCUGAUAUUUGACAGUUGAAAUAUCGAUCAUUUCAACUUGCA--- .....(((((....))))).((((((.......))).........(((((((((.......)))))))))))).--- ( -17.60, z-score = -1.55, R) >droPer1.super_28 432519 56 - 1111753 AGAAGAGGACAUUUGGUGUCGAGACACAGAUCGUGGCAUCUUUA-----GCCUAGUCCAUU---------------- ......((((.....((((....)))).......(((.......-----)))..))))...---------------- ( -14.40, z-score = -1.26, R) >dp4.chrXL_group1a 1987754 56 - 9151740 CGAAGAGGACAUUUGGUGUCGAGACACAGAUCGUGGCAUCUUUA-----GCCUAGUCCAUU---------------- ......((((.....((((....)))).......(((.......-----)))..))))...---------------- ( -14.40, z-score = -1.17, R) >droAna3.scaffold_13340 14430147 60 - 23697760 CGAAGAGGAUAUAUGGUUCUAUCACGAUGAUUGCCAUUACUGGG-----AAGAAAACAAGCAUUU------------ .....((.....(((((...(((.....))).)))))..))...-----................------------ ( -6.10, z-score = 0.89, R) >consensus ACCAGACGAUGUAUGGUGCCGCAACGAAGAGCGCGGCAUCUUUG_____AGCAAAUCAAGAAGUUCCACGAAU____ ..............((((((((..........))))))))..................................... ( -5.62 = -6.10 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:02 2011