
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,237,262 – 6,237,352 |
| Length | 90 |
| Max. P | 0.972031 |

| Location | 6,237,262 – 6,237,352 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Shannon entropy | 0.37256 |
| G+C content | 0.55162 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -18.54 |
| Energy contribution | -17.54 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
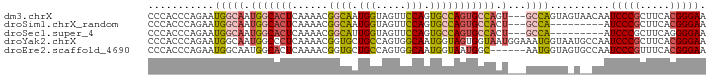
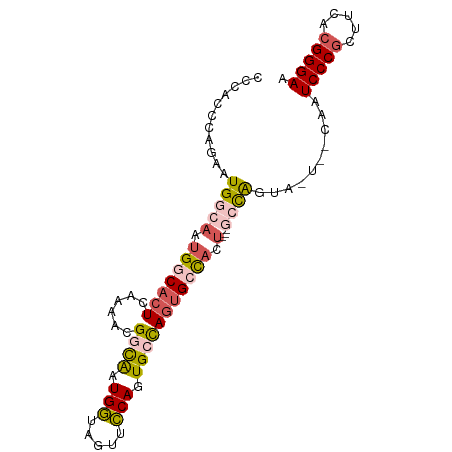
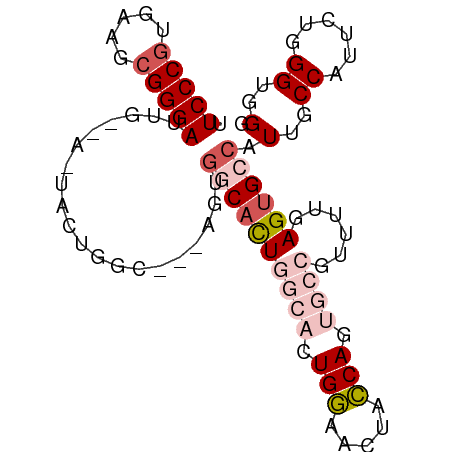
>dm3.chrX 6237262 90 + 22422827 CCCACCCAGAAUGGCAAUGGCACUCAAAACGGCAAUGGUAGUUCCAGUGCCAGUGCCAGU---GCCAGUAGUAACAAUCCCGCUUCACGGGAA ...........(((((.(((((((......((((.(((.....))).))))))))))).)---))))..........(((((.....))))). ( -36.00, z-score = -3.93, R) >droSim1.chrX_random 2021798 81 + 5698898 CCCACCCAGAAUGGCAAUGGCACUCAAAACGGCAAUGGUAGUUCCAGUGCCAGUGCCACU---GCCA---------AUCCCGCUUCACGGGAA ...........(((((.(((((((......((((.(((.....))).))))))))))).)---))))---------.(((((.....))))). ( -36.00, z-score = -4.39, R) >droSec1.super_4 5645876 81 - 6179234 CCCACCCAGAAUGGCAAUGGCACUCAAAACGGCAUUGGUAGUUCCAGUGCCAGUGCCACU---GCCA---------AUCCCGCUUCAGGGGAA ...........(((((.(((((((......((((((((.....))))))))))))))).)---))))---------.((((......)))).. ( -37.70, z-score = -4.29, R) >droYak2.chrX 2231691 93 + 21770863 CCCACCCAGAAUGGCAAUGGCCCUCAAAACGGUGCUGCCAGUGGCAAUGGUAGUGGUAAUGGAAAUGGUAAUGCCAAUCCCGCUUCACGGGAA ...........(((((...(((.((.....(.((((((((.......)))))))).)....))...)))..))))).(((((.....))))). ( -27.50, z-score = 0.71, R) >droEre2.scaffold_4690 15202300 87 + 18748788 CCCACCCAGAAUGGCAAUGGCACUCAAAACGGUGCUGCCAGUGGCAAUGGUAAUGGC------AAUGGUAGUGCCAAUCCCGUUUCACGGGAA ....(((....(((((...(((((......))))))))))((((.(((((...((((------(.......)))))...)))))))))))).. ( -32.70, z-score = -1.43, R) >consensus CCCACCCAGAAUGGCAAUGGCACUCAAAACGGCAAUGGUAGUUCCAGUGCCAGUGCCACU___GCCAGUA_U__CAAUCCCGCUUCACGGGAA .(((.((.....))...)))..........((((.(((((.......))))).))))....................(((((.....))))). (-18.54 = -17.54 + -1.00)
| Location | 6,237,262 – 6,237,352 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Shannon entropy | 0.37256 |
| G+C content | 0.55162 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
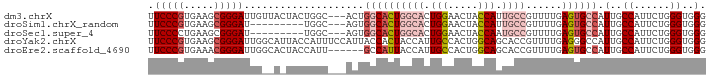
>dm3.chrX 6237262 90 - 22422827 UUCCCGUGAAGCGGGAUUGUUACUACUGGC---ACUGGCACUGGCACUGGAACUACCAUUGCCGUUUUGAGUGCCAUUGCCAUUCUGGGUGGG .(((((.....)))))...(((((..((((---(.(((((((((((.(((.....))).))))......))))))).))))).....))))). ( -38.10, z-score = -3.33, R) >droSim1.chrX_random 2021798 81 - 5698898 UUCCCGUGAAGCGGGAU---------UGGC---AGUGGCACUGGCACUGGAACUACCAUUGCCGUUUUGAGUGCCAUUGCCAUUCUGGGUGGG .(((((.....))))).---------((((---(((((((((((((.(((.....))).))))......)))))))))))))........... ( -39.80, z-score = -4.00, R) >droSec1.super_4 5645876 81 + 6179234 UUCCCCUGAAGCGGGAU---------UGGC---AGUGGCACUGGCACUGGAACUACCAAUGCCGUUUUGAGUGCCAUUGCCAUUCUGGGUGGG ...(((...(.(.(((.---------((((---(((((((((((((.(((.....))).))))......))))))))))))).))).).)))) ( -39.70, z-score = -3.86, R) >droYak2.chrX 2231691 93 - 21770863 UUCCCGUGAAGCGGGAUUGGCAUUACCAUUUCCAUUACCACUACCAUUGCCACUGGCAGCACCGUUUUGAGGGCCAUUGCCAUUCUGGGUGGG .(((((.....))))).(((.....))).........(((((.....((.((.((((..((......))...)))).)).)).....))))). ( -26.60, z-score = 0.37, R) >droEre2.scaffold_4690 15202300 87 - 18748788 UUCCCGUGAAACGGGAUUGGCACUACCAUU------GCCAUUACCAUUGCCACUGGCAGCACCGUUUUGAGUGCCAUUGCCAUUCUGGGUGGG .(((((.....))))).(((((.......)------))))...((((..(((.(((((((((........))))...)))))...))))))). ( -30.40, z-score = -1.30, R) >consensus UUCCCGUGAAGCGGGAUUG__A_UACUGGC___AGUGGCACUGGCACUGGAACUACCAGUGCCGUUUUGAGUGCCAUUGCCAUUCUGGGUGGG .(((((.....)))))....................((((((((((.(((.....))).))))......)))))).(..((......))..). (-19.66 = -21.62 + 1.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:22 2011