
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,214,741 – 6,214,852 |
| Length | 111 |
| Max. P | 0.554755 |

| Location | 6,214,741 – 6,214,852 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Shannon entropy | 0.52995 |
| G+C content | 0.63281 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -19.29 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
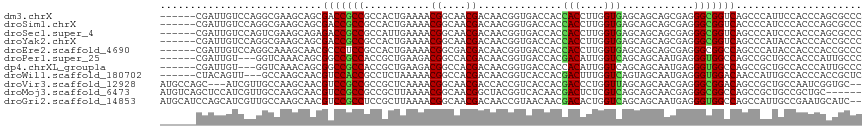
>dm3.chrX 6214741 111 + 22422827 ------CGAUUGUCCAGGCGAAGCAGCGACCGCCGCCACUGAAAACGGCAACGACAACGGUGACCACCACCUUGGUGAGCAGCAGCGAGGGCGGUCAGCCCAUUCCACCCAGCGCCC ------..........((((..((.((....(((((((.......((....)).....((((.....)))).))))).)).)).))..((((.....))))...........)))). ( -36.80, z-score = -0.16, R) >droSim1.chrX 4912048 111 + 17042790 ------CGAUUGUCCAGGCGAAGCAGCGACCGCCGCCACUGAAAACGGCAACGACAACGGUGACCACCACCUUGGUGAGCAGCAGCGAGGGCGGUCACCCCAUCCCACCCAGCGCCC ------..........((((..((.((....)).))..(((....((....)).....(((((((....(((((.((.....)).)))))..)))))))..........))))))). ( -37.40, z-score = -0.61, R) >droSec1.super_4 5625846 111 - 6179234 ------CGAUUGUCCAGUCGAAGCAGAGACCGCCGCCAUUGAAAACGGCAACGACAACGGUGACCACCACCUUGGUGAGCAGCAGCGAGGGCGGUCAGCCCAUCCCACCCAGCGCCC ------(((((....)))))..((...(((((((((((.......((....)).....((((.....)))).))))..((....))...))))))).((............)))).. ( -34.50, z-score = -0.51, R) >droYak2.chrX 2209626 111 + 21770863 ------CGAUUGUCCAGGCGAAGCAGCGACCGCCGCCACUGAAAACGGCAACGACAACGGUGACCACCACCUUGGUGAGCAGCAGCGAGGGCGGUCAGCCCAUACCACCCACCGCCC ------..........((((..((.((....(((((((.......((....)).....((((.....)))).))))).)).)).))..(((.(((........))).)))..)))). ( -37.20, z-score = -0.74, R) >droEre2.scaffold_4690 15181741 111 + 18748788 ------CGAUUGUCCAGGCAAAGCAACGCCCUCCGCCACUGAAAACGGCGACGACAACGGUGACCACCACCUUGGUGAGCAGCAGCGAGGGCGGCCAGCCCAUACCACCCACCGCCC ------...(((((..(((........)))...((((.........))))..)))))(((((..((((.....)))).((....))..((((.....))))........)))))... ( -35.80, z-score = -0.72, R) >droPer1.super_25 505566 108 + 1448063 ------CGAUUGU---GGUCAAACAGCGGCCGCCACCGCUGAAGACGGCCACGACAACGGUGACCACGACAUUGGUCAGCAGCAAUGAGGGUGGCCAGCCGCUGCCACCCAUUGCCC ------.(.((((---((((...((((((......)))))).....)))))))))...(.((((((......)))))).).(((((..(((((((........)))))))))))).. ( -48.30, z-score = -2.94, R) >dp4.chrXL_group1a 2347252 108 - 9151740 ------CGAUUGU---GGUCAAACAGCGGCCGCCACCGCUGAAGACGGCCACGACAACGGUGACCACCACAUUGGUCAGCAGCAAUGAGGGUGGCCAGCCGCUGCCACCCAUUGCCC ------.(.((((---((((...((((((......)))))).....)))))))))...(.((((((......)))))).).(((((..(((((((........)))))))))))).. ( -48.30, z-score = -2.96, R) >droWil1.scaffold_180702 3330786 108 - 4511350 ------CUACAGUU---GCCAAGCAACGUCCACCGCCUCUAAAAACGGCCACGACAACGGUCACCACGACUUUGGUCAGUAGCAAUGAGGGUGGACAACCAUUGCCACCCACCGCUC ------((((.(((---((...))))).......(((.........)))...(((...((((.....))))...))).))))......((((((.((.....))))))))....... ( -30.60, z-score = -1.18, R) >droVir3.scaffold_12928 7493361 112 + 7717345 AUGCCAGC---AUCGUUGCCAAGCAACGUCCGCCGCCGCUCAAAACGGCAACGACCACCGUCACCACGACCCUGGUUAGCAGCAACGAGGGCGGACAGCCGCUGCCAAUCGGUGC-- .((.((((---...(((((...)))))(((((((((((.......))))...(((((..(((.....)))..)))))............)))))))....)))).))........-- ( -42.60, z-score = -1.53, R) >droMoj3.scaffold_6473 14401755 111 - 16943266 AUGUCAGCUCCAUCGUUGCCAAGCAACGUCCGCCGCCGCUUAAAACGGCAACGGCUACGGUCACAACGACUCUCGUCAGCAGCAACGAGGGCGGCCAGCCGCUGCCGCUGC------ ....((((....(((((((...((..((((((..((((.......))))..)))..)))........(((....))).)).))))))).((((((.....)))))))))).------ ( -41.60, z-score = -0.90, R) >droGri2.scaffold_14853 491691 115 - 10151454 AUGCAUCCAGCAUCGUUGCCAAGCAACGUCCGCCUCCGCUUAAAACGGCAACGACAACCGUAACAACGACACUGGUCAGCAGCAAUGAGGGUGGCCAGCCAUUGCCGAAUGCAUC-- ((((((.(.(((((((((((((((.............)))).....))))))))....(((....)))...(((((((.((....))....)))))))....))).).)))))).-- ( -33.92, z-score = -0.61, R) >consensus ______CGAUUGUCCAGGCCAAGCAGCGACCGCCGCCACUGAAAACGGCAACGACAACGGUGACCACCACCUUGGUCAGCAGCAACGAGGGCGGCCAGCCCAUGCCACCCACCGCCC ...........................(((((((...........((....))..............(((....)))............)))))))..................... (-19.29 = -18.70 + -0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:16 2011