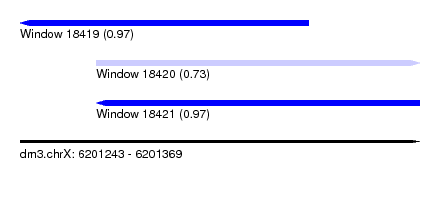
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,201,243 – 6,201,369 |
| Length | 126 |
| Max. P | 0.966980 |

| Location | 6,201,243 – 6,201,334 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 68.63 |
| Shannon entropy | 0.59174 |
| G+C content | 0.52843 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.67 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
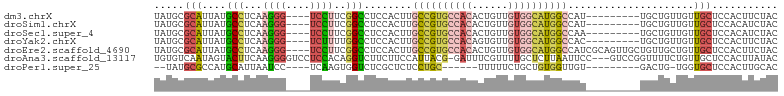
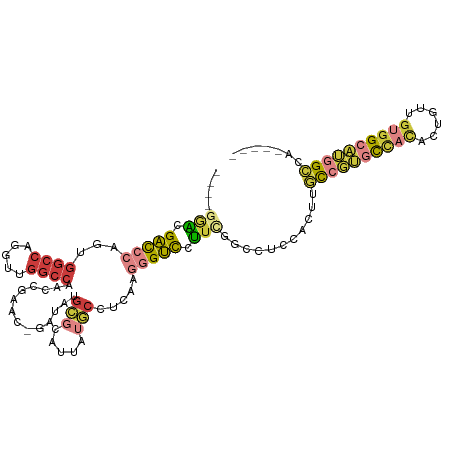
>dm3.chrX 6201243 91 - 22422827 UAUGCGCAUUAUGCCUCAAGGG----UCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAU---------UGCUGUUGUUGCUCCACUUCUAC .....(((....((..((((((----((....)))))......(((((((((((....)))))))))))..)---------))..))...)))........... ( -29.30, z-score = -1.54, R) >droSim1.chrX 4898963 91 - 17042790 UAUGCGCAUUAUGCCUCAAGGG----UCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAU---------UGCUGUUGUUGCUCCACAUCUAC .....(((....((..((((((----((....)))))......(((((((((((....)))))))))))..)---------))..))...)))........... ( -29.30, z-score = -1.26, R) >droSec1.super_4 5611487 91 + 6179234 UAUGCGCAUUAUGCCUCAAGGG----UCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAA---------UGCUGUUGUUGCUCCACAUCUAC .....(((((..(((..(((..----..))).)))........(((((((((((....))))))))))).))---------))).................... ( -30.10, z-score = -1.50, R) >droYak2.chrX 2196540 91 - 21770863 UAUGCGCAUUAUGCCUCAAGGG----UCUUUUGGCCUCCACUUGCCGUGCCACAGUGUUGUGGCAUGGCCAC---------UGCUGUUGUUGCUCCACUUCUAC ...(((((....((.....(((----((....)))))......((((((((((......))))))))))...---------.))...)).)))........... ( -29.90, z-score = -1.19, R) >droEre2.scaffold_4690 15168599 100 - 18748788 UAUGCGCAUUAUGCCUCAAGGG----UCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAUCGCAGUUGCUGUUGCUGUUGCUCCACUUCUAC .....(((...(((.....(((----((....)))))......(((((((((((....)))))))))))....)))..)))((..((....))..))....... ( -33.70, z-score = -1.48, R) >droAna3.scaffold_13117 2603090 100 + 5790199 UGUGUCAAUAGUACUUCAAGGGGUCCUCCACAGGUCUUCUUCCAUUACG-GAUUUCGUUUUGCUCUUAAUUCC---GUCCGGUUUUCUGUUGCUCCACUUAUAC .(((.((((((.((...((((((.(((....))).))))))......((-(((...(((........)))...---)))))))...))))))...)))...... ( -20.10, z-score = -1.28, R) >droPer1.super_25 486150 82 - 1448063 --UAUGCGCCAUGCAUUAAUCC----UCAAGUGGUCUCGCUCUCCUGC------UUUUUCUGCUGUGGUUGU---------GACUG-UGGUGCUCCACUUGCAC --...(((((((.(((....((----...((..(....((......))------.....)..))..))..))---------)...)-))))))........... ( -20.60, z-score = -0.96, R) >consensus UAUGCGCAUUAUGCCUCAAGGG____UCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAU_________UGCUGUUGUUGCUCCACUUCUAC .....(((....(((...((((....))))..)))........((((((((((......)))))))))).....................)))........... (-16.62 = -17.67 + 1.05)
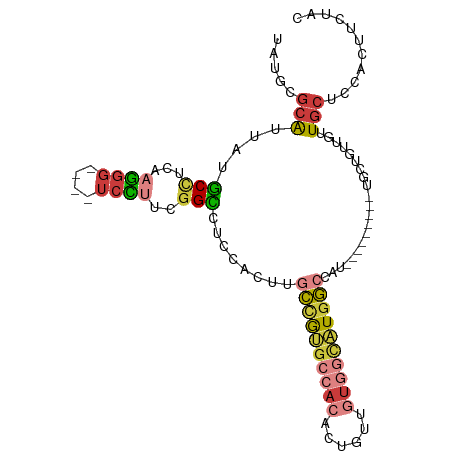
| Location | 6,201,267 – 6,201,369 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.58263 |
| G+C content | 0.57751 |
| Mean single sequence MFE | -36.29 |
| Consensus MFE | -20.10 |
| Energy contribution | -22.60 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
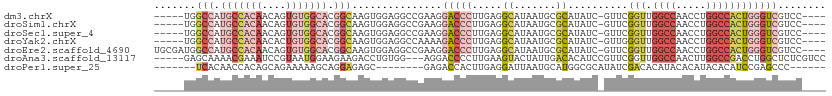
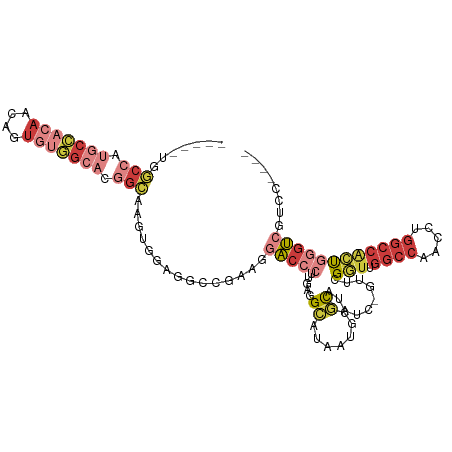
>dm3.chrX 6201267 102 + 22422827 -----UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUC-GUUCGGUUGGCCAACCUGGCCACUGGGUCGUCC---- -----..(((.(((((((....))))))).)))..(((.(.(((.(((....)))..)))....).)))....(-(((((((.((((.....)))))))))).))...---- ( -41.60, z-score = -0.99, R) >droSim1.chrX 4898987 102 + 17042790 -----UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUC-GUUCGGUUGGCCAACCUGGCCACUGGGUCGUCC---- -----..(((.(((((((....))))))).)))..(((.(.(((.(((....)))..)))....).)))....(-(((((((.((((.....)))))))))).))...---- ( -41.60, z-score = -0.99, R) >droSec1.super_4 5611511 102 - 6179234 -----UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUC-GUUCGGUUGGCCAACCUGGCCACUGGGUCGUCC---- -----..(((.(((((((....))))))).)))..(((.(.(((.(((....)))..)))....).)))....(-(((((((.((((.....)))))))))).))...---- ( -41.60, z-score = -0.99, R) >droYak2.chrX 2196564 102 + 21770863 -----UGGCCAUGCCACAACACUGUGGCACGGCAAGUGGAGGCCAAAAGACCCUUGAGGCAUAAUGCGCAUAUC-GUUGGGUUGGCCAACCUGGCCACUGGGUCGUCC---- -----..(((.(((((((....))))))).)))....(((((((....(((((.(((.((.......))...))-)..)))))((((.....))))....)))).)))---- ( -43.90, z-score = -1.84, R) >droEre2.scaffold_4690 15168627 107 + 18748788 UGCGAUGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUC-GUUCGGUUGGCCAACCUGGCCACUGGGUCGUCC---- ...(((((((.(((((((....))))))).(((.((.(..((((((.((((...((..((.....)).))....-))))..))))))..))).)))....))))))).---- ( -45.10, z-score = -1.35, R) >droAna3.scaffold_13117 2603126 104 - 5790199 -----GAGCAAAACGAAAUCCGUAAUGGAAGAAGACCUGUGG---AGGACCCCUUGAAGUACUAUUGACACAUCCGUUCGGUUGGCCAACUUGGCCGACCUGGCUCUCGUCC -----((((............(.((((((........(((((---(((...))))......(....).)))))))))))((((((((.....))))))))..))))...... ( -29.60, z-score = -0.72, R) >droPer1.super_25 486170 91 + 1448063 -------UCACAACCACAGCAGAAAAAGCAGGAGAGC--------GAGACCACUUGAGGAUUAAUGCAUGGCGCAUAUCGACACAUACACAUACACAUCCGAGCCC------ -------....................((.(((...(--------(((....))))..(((..((((.....)))))))..................)))..))..------ ( -10.60, z-score = 0.63, R) >consensus _____UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUC_GUUCGGUUGGCCAACCUGGCCACUGGGUCGUCC____ .......(((.(((((((....))))))).)))...............(((((.....((.......))..........(((.((((.....))))))))))))........ (-20.10 = -22.60 + 2.50)
| Location | 6,201,267 – 6,201,369 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.58263 |
| G+C content | 0.57751 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.71 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
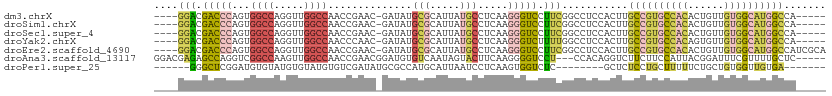
>dm3.chrX 6201267 102 - 22422827 ----GGACGACCCAGUGGCCAGGUUGGCCAACCGAAC-GAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA----- ----(((.(((((..(((((.....))))).......-.......((.....)).....)))))(....)..)))....(((((((((((....)))))))))))..----- ( -42.00, z-score = -1.70, R) >droSim1.chrX 4898987 102 - 17042790 ----GGACGACCCAGUGGCCAGGUUGGCCAACCGAAC-GAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA----- ----(((.(((((..(((((.....))))).......-.......((.....)).....)))))(....)..)))....(((((((((((....)))))))))))..----- ( -42.00, z-score = -1.70, R) >droSec1.super_4 5611511 102 + 6179234 ----GGACGACCCAGUGGCCAGGUUGGCCAACCGAAC-GAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA----- ----(((.(((((..(((((.....))))).......-.......((.....)).....)))))(....)..)))....(((((((((((....)))))))))))..----- ( -42.00, z-score = -1.70, R) >droYak2.chrX 2196564 102 - 21770863 ----GGACGACCCAGUGGCCAGGUUGGCCAACCCAAC-GAUAUGCGCAUUAUGCCUCAAGGGUCUUUUGGCCUCCACUUGCCGUGCCACAGUGUUGUGGCAUGGCCA----- ----....(((((..(((((.....))))).......-.......((.....)).....)))))...............((((((((((......))))))))))..----- ( -39.30, z-score = -0.77, R) >droEre2.scaffold_4690 15168627 107 - 18748788 ----GGACGACCCAGUGGCCAGGUUGGCCAACCGAAC-GAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAUCGCA ----(((.(((((..(((((.....))))).......-.......((.....)).....)))))(....)..)))....(((((((((((....)))))))))))....... ( -42.00, z-score = -1.12, R) >droAna3.scaffold_13117 2603126 104 + 5790199 GGACGAGAGCCAGGUCGGCCAAGUUGGCCAACCGAACGGAUGUGUCAAUAGUACUUCAAGGGGUCCU---CCACAGGUCUUCUUCCAUUACGGAUUUCGUUUUGCUC----- ......((((..(((.((((.....)))).)))(((((((...(((....(((....((((((.(((---....))).))))))....))).)))))))))).))))----- ( -32.30, z-score = -0.81, R) >droPer1.super_25 486170 91 - 1448063 ------GGGCUCGGAUGUGUAUGUGUAUGUGUCGAUAUGCGCCAUGCAUUAAUCCUCAAGUGGUCUC--------GCUCUCCUGCUUUUUCUGCUGUGGUUGUGA------- ------.((((.((((((((((((((((((....))))))).)))))))..))))......))))((--------((...((.((.......))...))..))))------- ( -23.80, z-score = -0.64, R) >consensus ____GGACGACCCAGUGGCCAGGUUGGCCAACCGAAC_GAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA_____ ....(((.(((((...((((.....))))..............(((.....))).....))))).)))...........((((((((((......))))))))))....... (-24.60 = -25.71 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:15 2011