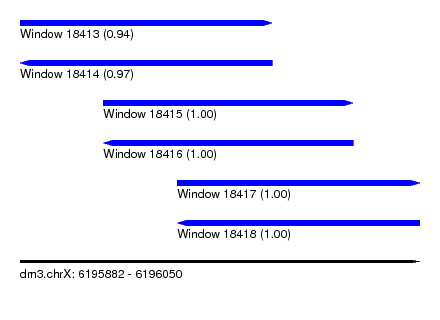
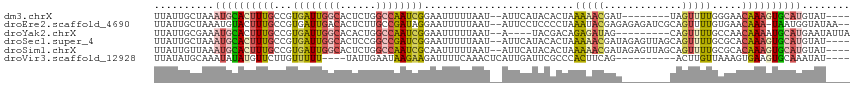
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,195,882 – 6,196,050 |
| Length | 168 |
| Max. P | 0.999229 |

| Location | 6,195,882 – 6,195,988 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.10 |
| Shannon entropy | 0.40331 |
| G+C content | 0.37139 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -19.08 |
| Energy contribution | -20.20 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
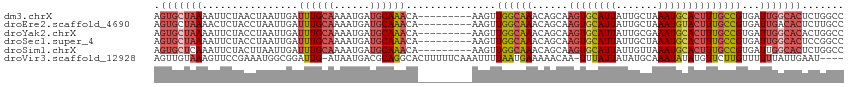
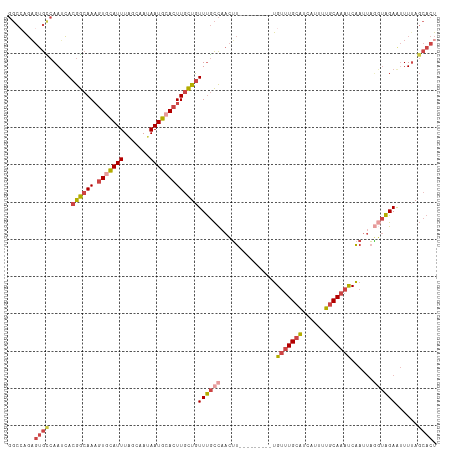
>dm3.chrX 6195882 106 + 22422827 AGUGCUAAAAUUCUAACUAAUUGAUUUGCAAAAUGAUGCAAACA---------AAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCC ((((((((....((((((..(((.((((((......))))))))---------)))))))...((.(((((((((((.......))))))).)))).))..))))))))...... ( -33.50, z-score = -3.04, R) >droEre2.scaffold_4690 15163228 106 + 18748788 AGUGCUAAAACUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACA---------AAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGUACUUUGCCGUGAUUGACACUCUUGCC ....................(((.((((((......))))))))---------)....(((((...(((((((((((.......))))))).)))).(((.....)))..))))) ( -23.20, z-score = -0.77, R) >droYak2.chrX 2191311 106 + 21770863 AGUGCUAAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACA---------AAGUUGGCAAACAGCAAGUGCAUUAUUGCGAAAUGCACUUUGCCGUGAUUGGCACACUGGCC .(((((((........((((((..((((((......))))))..---------.))))))...((.(((((((((((.......))))))).)))).))..)))))))....... ( -31.40, z-score = -1.94, R) >droSec1.super_4 5605964 106 - 6179234 AGUGCUAAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACA---------AAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCCGGCC ((((((((........((((((..((((((......))))))..---------.))))))...((.(((((((((((.......))))))).)))).))..))))))))...... ( -31.20, z-score = -2.17, R) >droSim1.chrX 4895783 106 + 17042790 AGUGCUCAAAUUCUACUUAAUUGAUUUGCAAAAUGAUGCAAACA---------AAGUUGGCAAACAGCAAGUGCAUUAUUGUUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCC (((((.(((..((.((....(((.((((((......))))))))---------)....(((((......((((((((.......)))))))))))))))))))))))))...... ( -28.70, z-score = -1.28, R) >droVir3.scaffold_12928 2732685 109 - 7717345 AGUUGUAAAGUUCCGAAAUGGCGGAUUG-AUAAUGACGCAGGCACUUUUUCAAAUUUUAAUGAAAAACAA-UUUAUUAUAUGCAAAUAUAUGUUCUUGUUUUUUAUUGAAU---- ..(((.(((((.((......(((.....-.......))).)).)))))..)))..(((((((((((((((-.....(((((....))))).....))))))))))))))).---- ( -22.80, z-score = -1.83, R) >consensus AGUGCUAAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACA_________AAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCC .(((((((................((((((......))))))...............((((((......((((((((.......))))))))))))))...)))))))....... (-19.08 = -20.20 + 1.12)
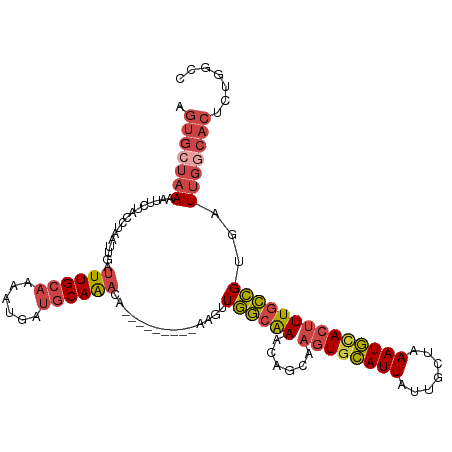
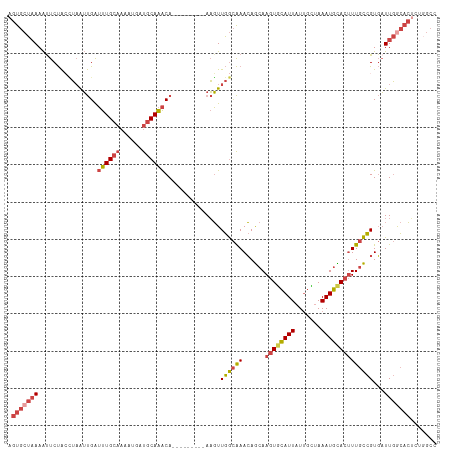
| Location | 6,195,882 – 6,195,988 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Shannon entropy | 0.40331 |
| G+C content | 0.37139 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6195882 106 - 22422827 GGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUU---------UGUUUGCAUCAUUUUGCAAAUCAAUUAGUUAGAAUUUUAGCACU ......(((((.....(((((((.(((((((.......)))))))))))))).....(((((---------((((((((......)))))).)))..)))).........))))) ( -31.50, z-score = -2.39, R) >droEre2.scaffold_4690 15163228 106 - 18748788 GGCAAGAGUGUCAAUCACGGCAAAGUACAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUU---------UGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAGUUUUAGCACU ......(((((......((((((.((.((((.......)))).))))))))((((((....(---------((((((((......)))))).)))...))))))......))))) ( -26.50, z-score = -1.07, R) >droYak2.chrX 2191311 106 - 21770863 GGCCAGUGUGCCAAUCACGGCAAAGUGCAUUUCGCAAUAAUGCACUUGCUGUUUGCCAACUU---------UGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUUAGCACU .......((((......((((((.(((((((.......)))))))))))))((((((....(---------((((((((......)))))).)))...))))))......)))). ( -34.30, z-score = -2.82, R) >droSec1.super_4 5605964 106 + 6179234 GGCCGGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUU---------UGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUUAGCACU ......(((((......((((((.(((((((.......)))))))))))))((((((....(---------((((((((......)))))).)))...))))))......))))) ( -35.10, z-score = -3.00, R) >droSim1.chrX 4895783 106 - 17042790 GGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAACAAUAAUGCACUUGCUGUUUGCCAACUU---------UGUUUGCAUCAUUUUGCAAAUCAAUUAAGUAGAAUUUGAGCACU ......((((((((...((((((.(((((((.......)))))))))))))(((((.....(---------((((((((......)))))).)))....)))))..))).))))) ( -33.40, z-score = -3.03, R) >droVir3.scaffold_12928 2732685 109 + 7717345 ----AUUCAAUAAAAAACAAGAACAUAUAUUUGCAUAUAAUAAA-UUGUUUUUCAUUAAAAUUUGAAAAAGUGCCUGCGUCAUUAU-CAAUCCGCCAUUUCGGAACUUUACAACU ----....(((.((((((((.....(((((....))))).....-)))))))).)))..........((((((....)........-...((((......)))))))))...... ( -10.90, z-score = 0.29, R) >consensus GGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUU_________UGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUUAGCACU .......((((.....(((((((.(((((((.......))))))))))))))....................(((((((......)))))))..................)))). (-19.84 = -20.77 + 0.92)

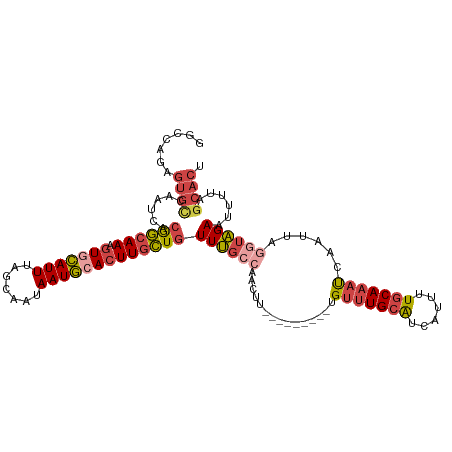
| Location | 6,195,917 – 6,196,022 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.52971 |
| G+C content | 0.36972 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -16.81 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.998889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
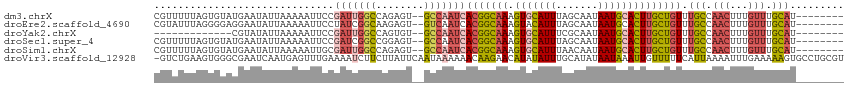
>dm3.chrX 6195917 105 + 22422827 --------AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGC--ACUCUGGCCAAUCGGAAUUUUUAAUAUUCAUACACUAAAAACG --------..((((.....((((.....)))).((((((((.......))))))))))))((((((((((--......))))))).((((.......))))...........))) ( -28.10, z-score = -2.61, R) >droEre2.scaffold_4690 15163263 105 + 18748788 --------AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGUACUUUGCCGUGAUUGAC--ACUCUUGCCGAUAGGAAUUUUUAAUAUUCCUCCCCUAAAUACG --------...........((((((((...(((((((((((.......))))))).)))).(((.....)--))..))))))))((((((.......))))))............ ( -26.00, z-score = -3.05, R) >droYak2.chrX 2191346 92 + 21770863 --------AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCGAAAUGCACUUUGCCGUGAUUGGC--ACACUGGCCAAUCGGAAUUUUUAAUAUACG------------- --------........((((((((((((..(((..((((....))))...)))..)))))).((((((((--......)))))))).)))))).........------------- ( -27.00, z-score = -2.07, R) >droSec1.super_4 5605999 105 - 6179234 --------AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGC--ACUCCGGCCGAUCGGAAUUUUUAAUAUUCAUACACUAAAAACG --------..((((.....((((.....)))).((((((((.......))))))))))))((((((((((--......))))))).((((.......))))...........))) ( -27.80, z-score = -2.22, R) >droSim1.chrX 4895818 105 + 17042790 --------AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGUUAAAUGCACUUUGCCGUGAUUGGC--ACUCUGGCCAAUCGCAAUUUUUAAUAUUCAUACACUAAAAACG --------..((((.....((((.....)))).((((((((.......)))))))))))).(((((((((--......)))))))))............................ ( -30.50, z-score = -3.41, R) >droVir3.scaffold_12928 2732719 114 - 7717345 ACGCAGGCACUUUUUCAAAUUUUAAUGAAAAACAAUUUAUUAUAUGCAAAUAUAUGUUCUUGUUUUUUAUUGAAUAAGAAGAUUUUCAAACUCAUUGAUUCGCCCACUUCAGAC- .....(((...(((((....(((((((((((((((.....(((((....))))).....)))))))))))))))...)))))...((((.....))))...)))..........- ( -22.00, z-score = -2.77, R) >consensus ________AUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGC__ACUCUGGCCAAUCGGAAUUUUUAAUAUUCAUACACUAAAAACG ..........(((((....((((.....))))..(((((((.......))))))))))))..((((((((........))))))))............................. (-16.81 = -16.40 + -0.41)
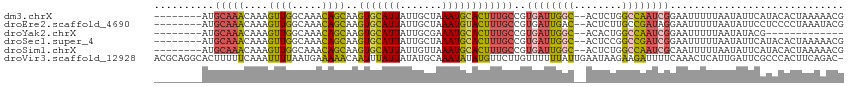
| Location | 6,195,917 – 6,196,022 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.52971 |
| G+C content | 0.36972 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
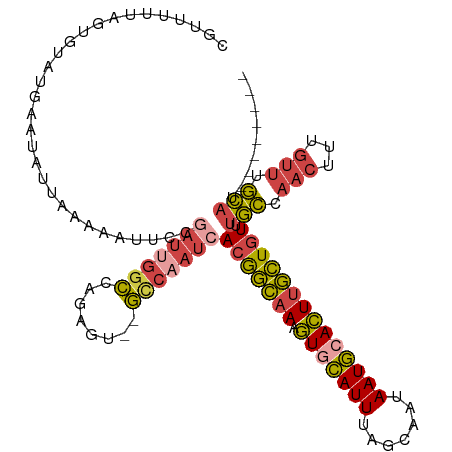
>dm3.chrX 6195917 105 - 22422827 CGUUUUUAGUGUAUGAAUAUUAAAAAUUCCGAUUGGCCAGAGU--GCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU-------- .(((((((((((....)))))))))))...(((((((......--)))))))(((((((.(((((((.......)))))))))))))).(((.(((...))).))).-------- ( -35.20, z-score = -3.94, R) >droEre2.scaffold_4690 15163263 105 - 18748788 CGUAUUUAGGGGAGGAAUAUUAAAAAUUCCUAUCGGCAAGAGU--GUCAAUCACGGCAAAGUACAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU-------- .(((..(((((.((((((.......))))))...((((((.((--(.....)))(((((.((.((((.......)))).))))))).))))))..)))))..)))..-------- ( -29.00, z-score = -2.52, R) >droYak2.chrX 2191346 92 - 21770863 -------------CGUAUAUUAAAAAUUCCGAUUGGCCAGUGU--GCCAAUCACGGCAAAGUGCAUUUCGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU-------- -------------.................(((((((......--)))))))(((((((.(((((((.......)))))))))))))).(((.(((...))).))).-------- ( -29.70, z-score = -2.99, R) >droSec1.super_4 5605999 105 + 6179234 CGUUUUUAGUGUAUGAAUAUUAAAAAUUCCGAUCGGCCGGAGU--GCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU-------- .(((((((((((....)))))))))))...(((.(((......--))).)))(((((((.(((((((.......)))))))))))))).(((.(((...))).))).-------- ( -31.10, z-score = -2.31, R) >droSim1.chrX 4895818 105 - 17042790 CGUUUUUAGUGUAUGAAUAUUAAAAAUUGCGAUUGGCCAGAGU--GCCAAUCACGGCAAAGUGCAUUUAACAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU-------- .(((((((((((....)))))))))))((((((((((......--)))))))(((((((.(((((((.......))))))))))))))...............))).-------- ( -36.00, z-score = -3.97, R) >droVir3.scaffold_12928 2732719 114 + 7717345 -GUCUGAAGUGGGCGAAUCAAUGAGUUUGAAAAUCUUCUUAUUCAAUAAAAAACAAGAACAUAUAUUUGCAUAUAAUAAAUUGUUUUUCAUUAAAAUUUGAAAAAGUGCCUGCGU -.......(..((((..(((((((((..(((....)))..)))))...((((((((.....(((((....))))).....)))))))).........)))).....))))..).. ( -21.70, z-score = -1.63, R) >consensus CGUUUUUAGUGUAUGAAUAUUAAAAAUUCCGAUUGGCCAGAGU__GCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAU________ ..............................(((((((........)))))))(((((((.(((((((.......)))))))))))))).(((.(((...))).)))......... (-18.40 = -18.88 + 0.48)
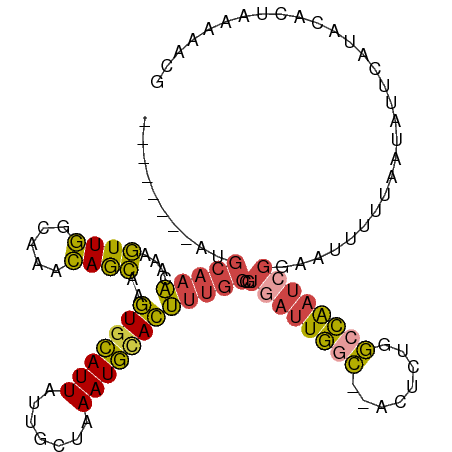
| Location | 6,195,948 – 6,196,050 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.60019 |
| G+C content | 0.35176 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6195948 102 + 22422827 UUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUAAU--AUUCAUACACUAAAAACGAU--------UAGUUUUGGGAACAAAGUGCAUGUAU---- ..........((((((((((..(((((((((......))))))).((((.......--))))..)).((.(((((...--------..))))).))..))))))))))....---- ( -29.10, z-score = -3.58, R) >droEre2.scaffold_4690 15163294 111 + 18748788 UUAUUGCUAAAUGUACUUUGCCGUGAUUGACACUCUUGCCGAUAGGAAUUUUUAAU--AUUCCUCCCCUAAAUACGAGAGAGAUCGCAGUUUUGUGAACAAA-UAAUGGUAUAA-- ....(((.....)))...((((((((((....((((((.....((((((.......--))))))..........)))))).))))))...((((....))))-....))))...-- ( -21.96, z-score = -1.57, R) >droYak2.chrX 2191377 101 + 21770863 UUAUUGCGAAAUGCACUUUGCCGUGAUUGGCACACUGGCCAAUCGGAAUUUUUAAU--A----UACGACAGAGAUAG---------CAGUUUUGCCAACAAAAUGCAUGAAUAUUA .....(((((((((.(((((.((((((((((......)))))))............--.----.))).)))))...)---------)).))))))..................... ( -22.53, z-score = -1.12, R) >droSec1.super_4 5606030 110 - 6179234 UUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCCGGCCGAUCGGAAUUUUUAAU--AUUCAUACACUAAAAACGAUAGAGUUAGCAGUUUUGCGCACAAAGUGCAUGUAU---- ..(((((((.....((((((.((((((((((......))))))).((((.......--))))...........))).)))))))))))))..(((((.....))))).....---- ( -29.50, z-score = -1.98, R) >droSim1.chrX 4895849 110 + 17042790 UUAUUGUUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGCAAUUUUUAAU--AUUCAUACACUAAAAACGAUAGAGUUAGCAGUUUUGCGCACAAAGUGCAUGUAU---- ..........((((((((((..(((((((((......)))))))))..........--...........................((......))...))))))))))....---- ( -30.50, z-score = -2.53, R) >droVir3.scaffold_12928 2732758 98 - 7717345 UUAUAUGCAAAUAUAUGUUCUUGUUUUU----UAUUGAAUAAGAAGAUUUUCAAACUCAUUGAUUCGCCCACUUCAG----------ACUUGUUAAAGUGAAGUGCAAAUAU---- ......((.........(((((((((..----....)))))))))((...((((.....)))).)))).((((((..----------((((....)))))))))).......---- ( -18.20, z-score = -1.51, R) >consensus UUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUAAU__AUUCAUACACUAAAAACGAU________CAGUUUUGCGAACAAAGUGCAUGUAU____ ..........((((((((((...((((((((......)))))))).........................(((((.............))))).....))))))))))........ (-13.80 = -14.75 + 0.95)
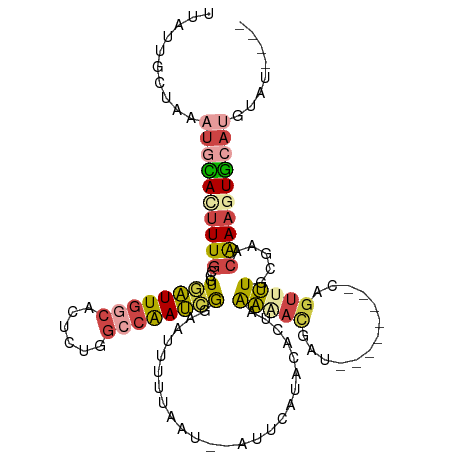
| Location | 6,195,948 – 6,196,050 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.60019 |
| G+C content | 0.35176 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -11.17 |
| Energy contribution | -13.15 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6195948 102 - 22422827 ----AUACAUGCACUUUGUUCCCAAAACUA--------AUCGUUUUUAGUGUAUGAAU--AUUAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAA ----.((.((((((((((((..........--------...(((((((((((....))--)))))))))...(((((((......)))))))..)))))))))))).))....... ( -31.50, z-score = -5.09, R) >droEre2.scaffold_4690 15163294 111 - 18748788 --UUAUACCAUUA-UUUGUUCACAAAACUGCGAUCUCUCUCGUAUUUAGGGGAGGAAU--AUUAAAAAUUCCUAUCGGCAAGAGUGUCAAUCACGGCAAAGUACAUUUAGCAAUAA --...........-((((....))))..(((((......))((((((..(..((((((--.......))))))..).((....(((.....))).)).)))))).....))).... ( -17.80, z-score = -0.22, R) >droYak2.chrX 2191377 101 - 21770863 UAAUAUUCAUGCAUUUUGUUGGCAAAACUG---------CUAUCUCUGUCGUA----U--AUUAAAAAUUCCGAUUGGCCAGUGUGCCAAUCACGGCAAAGUGCAUUUCGCAAUAA ........((((((((((((((((....))---------).............----.--............(((((((......))))))).))))))))))))).......... ( -28.30, z-score = -3.13, R) >droSec1.super_4 5606030 110 + 6179234 ----AUACAUGCACUUUGUGCGCAAAACUGCUAACUCUAUCGUUUUUAGUGUAUGAAU--AUUAAAAAUUCCGAUCGGCCGGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAA ----.((.(((((((((((..(((....))).........((((((((((((....))--)))))))((((((......)))))).......)))))))))))))).))....... ( -29.30, z-score = -2.02, R) >droSim1.chrX 4895849 110 - 17042790 ----AUACAUGCACUUUGUGCGCAAAACUGCUAACUCUAUCGUUUUUAGUGUAUGAAU--AUUAAAAAUUGCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAACAAUAA ----.((.((((((((((((((((....)))..........(((((((((((....))--))))))))).))(((((((......)))))))...))))))))))).))....... ( -35.80, z-score = -4.31, R) >droVir3.scaffold_12928 2732758 98 + 7717345 ----AUAUUUGCACUUCACUUUAACAAGU----------CUGAAGUGGGCGAAUCAAUGAGUUUGAAAAUCUUCUUAUUCAAUA----AAAAACAAGAACAUAUAUUUGCAUAUAA ----.(((.((((.(((((((((......----------.)))))))))........(((((..(((....)))..)))))...----...................)))).))). ( -16.00, z-score = -1.10, R) >consensus ____AUACAUGCACUUUGUUCGCAAAACUG________AUCGUUUUUAGUGUAUGAAU__AUUAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAA .........((((((((((.........................((((((..........))))))......(((((((......)))))))...))))))))))........... (-11.17 = -13.15 + 1.98)

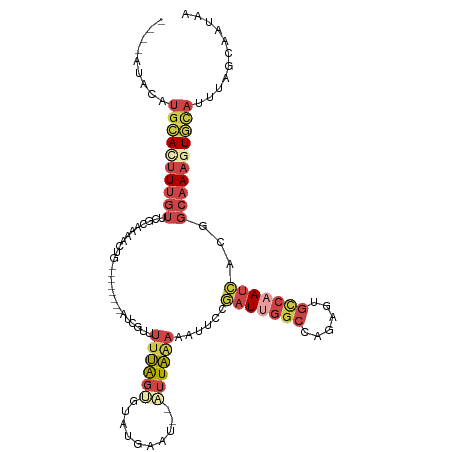
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:13 2011