| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,188,593 – 6,188,759 |
| Length | 166 |
| Max. P | 0.623312 |

| Location | 6,188,593 – 6,188,759 |
|---|---|
| Length | 166 |
| Sequences | 10 |
| Columns | 184 |
| Reading direction | reverse |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.73160 |
| G+C content | 0.50436 |
| Mean single sequence MFE | -50.09 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
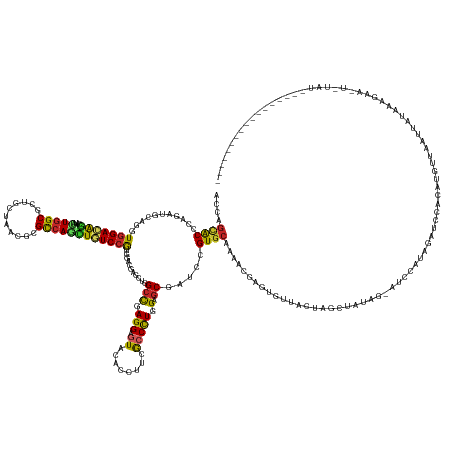
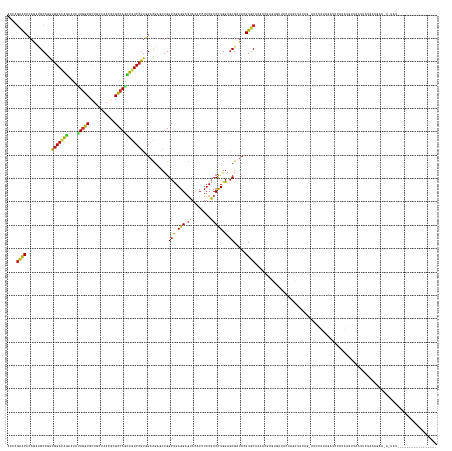
>dm3.chrX 6188593 166 - 22422827 ACCAGCACCCAAAUGCAGGUGGACAGCAUAUUGGCGCUACUAACGCGCCAAUUGUCCGUGGACGAGCUGGCCGAGGAGUACACCUUCGCCCUGGAGCGAUCCGUGCAGAACGAGUGUUACUAGCUUUAG-AUCCAUAGAUCCACUGGUUAAUAUUCAUACAUUACAU----------------- ....((((............(((((....((((((((.......)))))))))))))..(((((..(((((.((((......)))).)))..))..)).))))))).....((((((((((((.....(-(((....))))..)))).))))))))...........----------------- ( -53.80, z-score = -2.15, R) >droSim1.chrX 4891273 164 - 17042790 ACCAGCACCCAGAUGCAGGUGGACAGCAUACUGGCGCUACUAACACGCCAGCUGUCCGUGGACGAGCUGGCCGAGGAGUACACCUUCGCCCUGGAGCGAUCCGUGCAGAACGAGUGUUACUAGCUUUAG--UCCACAGAUCCACUGGUUACUAUACAUGCA-UACAU----------------- ....(((.((((.((...((((((.((......))((((.((((((((((((((((....))).)))))))......((((....((((......))))...)))).......)))))).))))....)--))))).....))))))..........))).-.....----------------- ( -56.70, z-score = -1.47, R) >droSec1.super_4 5598797 164 + 6179234 ACCAGCACCCAGAUGCAGGUGGACAGCAUAUUGGCGCUACUAACGCGCCAGCUGUCCGUGGACGAGCUGGCCGAGGAGUACACCUUCGCCCUGGAGCGAUCCGUGCAGAACGAGUGUUACUAGCUUUAG--UCCACAGAUCCACUGGUUAAUAUACAUGCA-UACAU----------------- (((((.(((........)))(((((((....((((((.......)))))))))))))((((((((((((((.((((......)))).)))..((((((...(((.....)))..)))).)))))))..)--))))).......))))).............-.....----------------- ( -58.30, z-score = -1.90, R) >droEre2.scaffold_4690 15155890 152 - 18748788 ACCAGCACCCAAAUGCAGGUGGACAGCAUCCUGGCGCUGCUCACGCGCCAACUGUCCGUGGACGAGCUGGCCGAGGAGUAUACCUUCGCCCUAGAGCGAUCCGUGCAAAACGAGUGUUACUAGCAUUAG--UCCGUAGAUCCACUGGUUAAUAU------------------------------ (((.(((......))).)))((((((.....((((((.......)))))).))))))(((((((..(((((.((((......)))).)))((((((((...(((.....)))..)))).))))....))--..))....)))))..........------------------------------ ( -50.30, z-score = -0.84, R) >droAna3.scaffold_13117 1748789 137 - 5790199 GCCAGCACCCAGAUGCAAGUGGACAGCAUUCUGGCGCUGCUCACACGUCAAAUGUCCAUGGACGAACUGGCUGAGGAGUACACGUUCGCCUUGGAACGCUCUGUCCAGAACGAGUCUUAAUAGUUAAAA-AUUCCCUG---------------------------------------------- (((((......((((...(((..((((........))))..))).))))...((((....))))..)))))...(((((...(((((......)))))..((((..(((.....)))..))))......-)))))...---------------------------------------------- ( -37.70, z-score = 0.07, R) >dp4.chrXL_group3a 491379 181 + 2690836 CUCAGUGCCGAGCAGCAGAUGGACGCCGCCUUGGCCCUCUUAACGCGCCAGGUGUCCGUCGAUCGUCUGGCCGAGGAAUACACCUUCGCCCUGGAGCGGUCCGUGCAAAAGGAUAGCAACUAG--AGAG-CUGAUUAGAUGUAUAUUUUAACUACAAAUAGCUUUAUCAUAGUAUUAAUGUAAU ....((.((.....(((((((((((((....((((.(.......).))))))))))))))(((((((((((.((((......)))).)))..))).)))))..)))....))...)).((((.--((((-(((..(((.((.......)).)))....)))))))....))))........... ( -52.60, z-score = -0.24, R) >droPer1.super_11 2476095 181 - 2846995 CUCAGUGCCGAGCAGCAGAUGGACGCCGCCUUGGCCCUCUUAACGCGCCAGGUGUCCGUCGAUCGUCUGGCCGAGGAAUACACCUUCGCCCUGGAGCGGUCCGUGCAAAAGGAUAGCAACUAG--AGAG-CUGAUUAGAUGUAUAUUUUAACUACAAAUAGCUUUAUCAUAGUUUUAAUGUCAU ((...(((((.......((((((((((....((((.(.......).))))))))))))))(((((((((((.((((......)))).)))..))).))))))).)))..))((((..(((((.--((((-(((..(((.((.......)).)))....)))))))....)))))....)))).. ( -55.10, z-score = -0.76, R) >droVir3.scaffold_12932 1332975 169 - 2102469 AAUGGCACCCAGUUGCAAGUGGACGGCAUUUUAGCACUGAUGACCCGUCAGCUCUCGGUGGAUGCACUGGCCGAGGAGUAUACAUUCACCCUGGAGCGUUCAGUGCAAAAUGAUUCGUACUGAAUAUGGCGAGCAAUGAAACUAAUGCUAAUUGUAUACAUAAUUUAAU--------------- ..(((((....(((((........((((((........)))).))((((((((((.((((((((.(((........)))...))))))))..)))))(((((((((..........))))))))).))))).)))))........)))))...................--------------- ( -47.50, z-score = -0.45, R) >droMoj3.scaffold_6473 11708642 169 + 16943266 AAUUACACACAGACGCAAGUGGAUGGUAUUCUGGCAUUGAUGACGCGCCAGCUAUCCGUCGAUGCCAUGGCCGAGGAGUACACAUUCGCCCUGGAGCGUUCAGUGCAAAAUGAUCCCUACUAAAAGGGCGACCAUGGGAACCUAAUGCUUAUUAGAUAUAUAUCUAUAU--------------- .........(((((....))((((((((((((((((((((((..((....))....))))))))))........))))))).)))))...)))(((((((..((.(...(((.(((((.......))).)).)))..).))..)))))))..(((((....)))))...--------------- ( -47.90, z-score = -0.62, R) >droGri2.scaffold_14853 7078111 162 - 10151454 AGUGGCACGCAGAUGCAAGUGGACGGCAUAUUGGCUUUUCUGACGCGUCAGCUAUCCGUGGAUGCACUGGCCGAGGAGUACACAUUCACCCUGGAGCGUUCAGUGCAAAAUGAGUCCUACUGAAUCAAUGACAAAAUGAAGCCUAUGCUAAUUAUAUAGAAU---------------------- ..(((((.((.(((...(((((((..(((....((....((((((((((((.((((....))))..)))))(.(((.((........))))).).))).)))).))...))).)))).)))..))).....(.....)..))...)))))............---------------------- ( -41.00, z-score = 1.13, R) >consensus ACCAGCACCCAGAUGCAGGUGGACAGCAUAUUGGCGCUGCUAACGCGCCAGCUGUCCGUGGACGAGCUGGCCGAGGAGUACACCUUCGCCCUGGAGCGAUCCGUGCAAAACGAGUGUUACUAGCUAUAG_AUCCAUAGAUCCACAUGUUAAUUAUAAAGAA_U_UAU_________________ ....((((...........(((((((....(((((...........))))))))))))...........(((.(((.((........))))).).)).....)))).............................................................................. (-21.66 = -21.00 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:06 2011