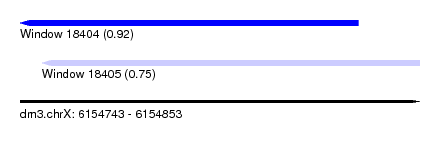
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,154,743 – 6,154,853 |
| Length | 110 |
| Max. P | 0.923587 |

| Location | 6,154,743 – 6,154,836 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.96 |
| Shannon entropy | 0.22385 |
| G+C content | 0.39246 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
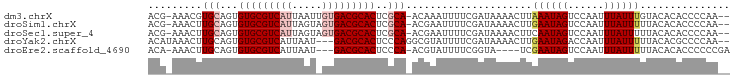
>dm3.chrX 6154743 93 - 22422827 ACG-AAACGUGCAGUGUGCGUCAUUAAUUGUGACGCACUCGCA-ACAAAUUUUCGAUAAAACUUAAAUAGUCCAAUUUAUUUGUACACACCCCAA-- .((-(((..(((.(.(((((((((.....))))))))).))))-......)))))........(((((((......)))))))............-- ( -23.20, z-score = -3.21, R) >droSim1.chrX 4865090 93 - 17042790 ACG-AAACUUGCAGUGUGCGUCAUUAGUAGUGACGCACUCGCA-ACGAAUUUUCGAUAAAACUUGAAUAGUCCAAUUUAUUUUUACACACCCCAA-- .((-((((((((.(.(((((((((.....))))))))).))))-).)...))))).(((((..(((((......))))).)))))..........-- ( -22.20, z-score = -2.91, R) >droSec1.super_4 5564110 93 + 6179234 ACG-AAACUUGCAGUGUGCGUCAUUAGUAGUGACGCACUCGCA-ACGAAUUUUCGAUAAAACUUCAAUAGUCCAAUUUAUUUUUACACACCCCAA-- .((-((((((((.(.(((((((((.....))))))))).))))-).)...)))))........................................-- ( -21.30, z-score = -3.06, R) >droYak2.chrX 2158758 92 - 21770863 ACAUAAACUUGCAGUGUGCGUCAUUAAU---GACGCACUCCCAGGCGUAUUUUCGAUAAAACUUGAAUAGACCAAUUUAUUUUUACACGCCCCAA-- .............(.((((((((....)---))))))).)...(((((......((((((..(((.......))))))))).....)))))....-- ( -20.90, z-score = -3.07, R) >droEre2.scaffold_4690 15125021 88 - 18748788 ACA-AAACUUGCAGUGUGCGUCAUUAAU---GACGCACUCCCA-ACGUAUUUUCGGUA----UCGAAUAGUCCAAUUUAUUUUUACACACCCCCCGA ...-..(((((..(.((((((((....)---))))))).).))-).)).......(((----..((((((......)))))).)))........... ( -16.50, z-score = -1.91, R) >consensus ACG_AAACUUGCAGUGUGCGUCAUUAAU_GUGACGCACUCGCA_ACGAAUUUUCGAUAAAACUUGAAUAGUCCAAUUUAUUUUUACACACCCCAA__ .............(.(((((((((.....))))))))).)........................((((((......))))))............... (-12.84 = -13.72 + 0.88)
| Location | 6,154,749 – 6,154,853 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.14 |
| Shannon entropy | 0.27553 |
| G+C content | 0.35937 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -14.08 |
| Energy contribution | -14.56 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
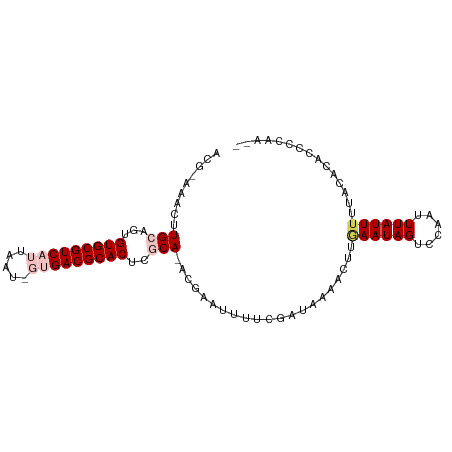
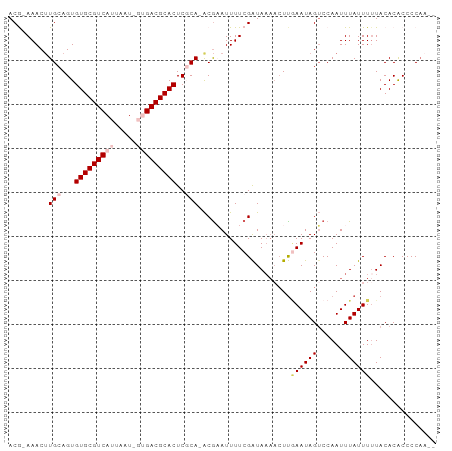
>dm3.chrX 6154749 104 - 22422827 GACCUUUAUU--GCUGAUAACG-AAACGUGCAGUGUGCGUCAUUAAUUGUGACGCACUCGCA-ACAAAUUUUCGAUAAAACUUAAAUAGUCCAAUUUAUUUGUACACA .........(--((.(((((((-(((..(((.(.(((((((((.....))))))))).))))-......))))).....(((.....))).....))))).))).... ( -23.60, z-score = -1.91, R) >droSim1.chrX 4865096 104 - 17042790 GACCUUUAUU--GCUGAUAACG-AAACUUGCAGUGUGCGUCAUUAGUAGUGACGCACUCGCA-ACGAAUUUUCGAUAAAACUUGAAUAGUCCAAUUUAUUUUUACACA ....((((((--(..(((..((-....((((.(.(((((((((.....))))))))).))))-))).)))..)))))))....((((((......))))))....... ( -22.90, z-score = -1.71, R) >droSec1.super_4 5564116 99 + 6179234 -----GUUUU--UUUGAUAACG-AAACUUGCAGUGUGCGUCAUUAGUAGUGACGCACUCGCA-ACGAAUUUUCGAUAAAACUUCAAUAGUCCAAUUUAUUUUUACACA -----(((((--.((((.....-....((((.(.(((((((((.....))))))))).))))-).......)))).)))))........................... ( -23.49, z-score = -2.56, R) >droYak2.chrX 2158764 105 - 21770863 GACCUUUGUUCAGUUGAUAACAUAAACUUGCAGUGUGCGUCAUUAA---UGACGCACUCCCAGGCGUAUUUUCGAUAAAACUUGAAUAGACCAAUUUAUUUUUACACG ....(((((((((((((....(((..((((..(.((((((((....---)))))))).).))))..)))..)))))......)))))))).................. ( -23.40, z-score = -2.44, R) >droEre2.scaffold_4690 15125029 97 - 18748788 GACCUUUAUC--ACUUAUAACA-AAACUUGCAGUGUGCGUCAUUAA---UGACGCACUCCCA-ACGUAUUUUCGGUA----UCGAAUAGUCCAAUUUAUUUUUACACA (((.......--..........-..(((((..(.((((((((....---)))))))).).))-).))...((((...----.))))..)))................. ( -17.00, z-score = -1.99, R) >consensus GACCUUUAUU__GCUGAUAACG_AAACUUGCAGUGUGCGUCAUUAAU_GUGACGCACUCGCA_ACGAAUUUUCGAUAAAACUUGAAUAGUCCAAUUUAUUUUUACACA ................................(.(((((((((.....))))))))).)........................((((((......))))))....... (-14.08 = -14.56 + 0.48)

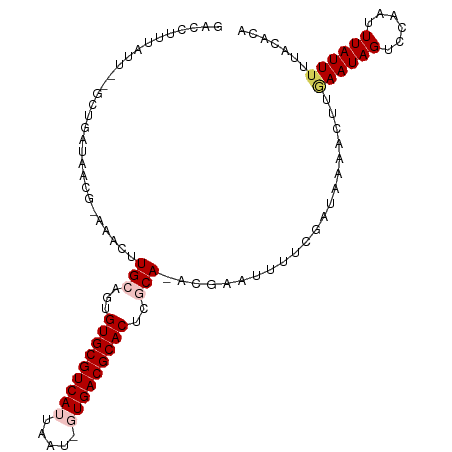
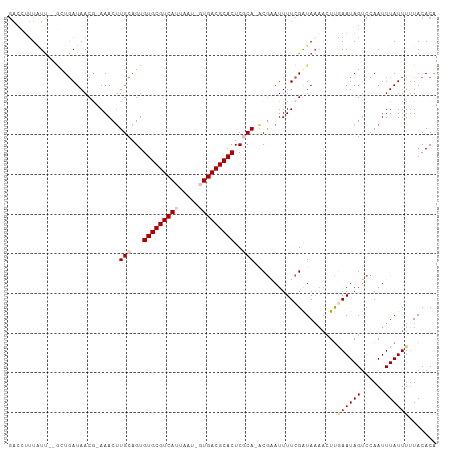
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:20:02 2011